 Found 4 hits Enzyme Inhibition Constant Data
Found 4 hits Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Histone-lysine N-methyltransferase EHMT1
(285/285 = 100%)†
(Homo sapiens (Human)) | BDBM50396024
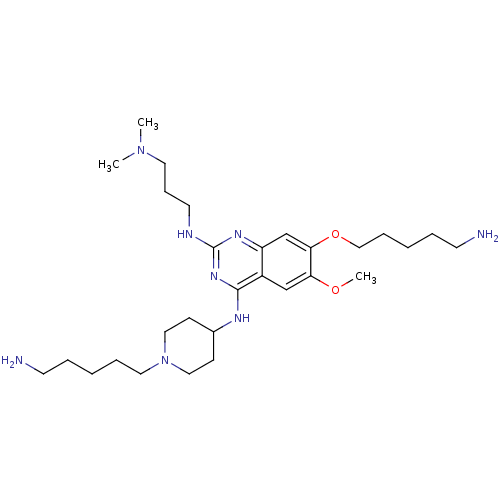
(CHEMBL1232453)Show SMILES COc1cc2c(NC3CCN(CCCCCN)CC3)nc(NCCCN(C)C)nc2cc1OCCCCCN Show InChI InChI=1S/C29H52N8O2/c1-36(2)16-10-15-32-29-34-25-22-27(39-20-9-5-7-14-31)26(38-3)21-24(25)28(35-29)33-23-11-18-37(19-12-23)17-8-4-6-13-30/h21-23H,4-20,30-31H2,1-3H3,(H2,32,33,34,35) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 100 | n/a | n/a | n/a | n/a | n/a | n/a |
Icahn School of Medicine at Mount Sinai
Curated by ChEMBL
| Assay Description
Inhibition of recombinant human GLP (951 to 1235) using Histone H3 peptide (1 to 15) as substrate preincubated on ice for 10 mins followed by 5 mins ... |
J Med Chem 58: 1596-629 (2015)
Article DOI: 10.1021/jm501234a
BindingDB Entry DOI: 10.7270/Q28K7BS2 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(285/285 = 100%)†
(Homo sapiens (Human)) | BDBM50009672
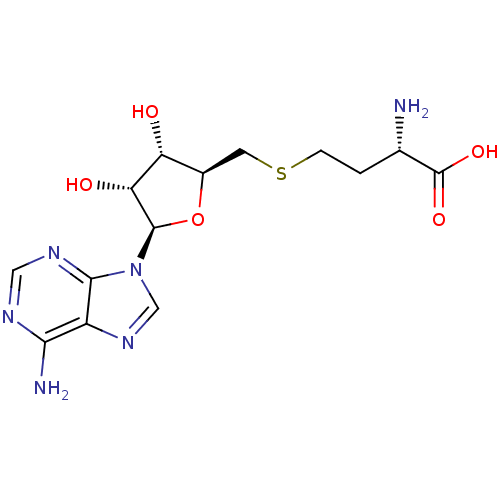
(AdoHcy | CHEMBL418052 | S-(5'-adenosyl)-L-homocyst...)Show SMILES N[C@@H](CCSC[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(N)ncnc12)C(O)=O Show InChI InChI=1S/C14H20N6O5S/c15-6(14(23)24)1-2-26-3-7-9(21)10(22)13(25-7)20-5-19-8-11(16)17-4-18-12(8)20/h4-7,9-10,13,21-22H,1-3,15H2,(H,23,24)(H2,16,17,18)/t6-,7+,9+,10+,13+/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
KEGG
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 230 | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc.
Curated by ChEMBL
| Assay Description
Inhibition of EHMT1 (unknown origin) by HMT assay |
Bioorg Med Chem Lett 25: 1532-7 (2015)
Article DOI: 10.1016/j.bmcl.2015.02.017
BindingDB Entry DOI: 10.7270/Q2TM7CSS |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(285/285 = 100%)†
(Homo sapiens (Human)) | BDBM50396024

(CHEMBL1232453)Show SMILES COc1cc2c(NC3CCN(CCCCCN)CC3)nc(NCCCN(C)C)nc2cc1OCCCCCN Show InChI InChI=1S/C29H52N8O2/c1-36(2)16-10-15-32-29-34-25-22-27(39-20-9-5-7-14-31)26(38-3)21-24(25)28(35-29)33-23-11-18-37(19-12-23)17-8-4-6-13-30/h21-23H,4-20,30-31H2,1-3H3,(H2,32,33,34,35) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 136 | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Binding affinity to human recombinant GLP catalytic domain amino acid 951 to 1235 expressed in Escherichia coli BL21 (DE3) by isothermal titration ca... |
Eur J Med Chem 56: 179-194 (2012)
Article DOI: 10.1016/j.ejmech.2012.08.010
BindingDB Entry DOI: 10.7270/Q2TQ62NX |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(285/285 = 100%)†
(Homo sapiens (Human)) | BDBM50396024

(CHEMBL1232453)Show SMILES COc1cc2c(NC3CCN(CCCCCN)CC3)nc(NCCCN(C)C)nc2cc1OCCCCCN Show InChI InChI=1S/C29H52N8O2/c1-36(2)16-10-15-32-29-34-25-22-27(39-20-9-5-7-14-31)26(38-3)21-24(25)28(35-29)33-23-11-18-37(19-12-23)17-8-4-6-13-30/h21-23H,4-20,30-31H2,1-3H3,(H2,32,33,34,35) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 136 | n/a | n/a | n/a | n/a | n/a |
Icahn School of Medicine at Mount Sinai
Curated by ChEMBL
| Assay Description
Binding affinity to recombinant human GLP (951 to 1235) by isothermal titration calorimetric analysis |
J Med Chem 58: 1596-629 (2015)
Article DOI: 10.1021/jm501234a
BindingDB Entry DOI: 10.7270/Q28K7BS2 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data