

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyclin-A1/Cyclin-A2/Cyclin-dependent kinase 2 (253/262 = 97%)† (Homo sapiens (Human)) | BDBM11324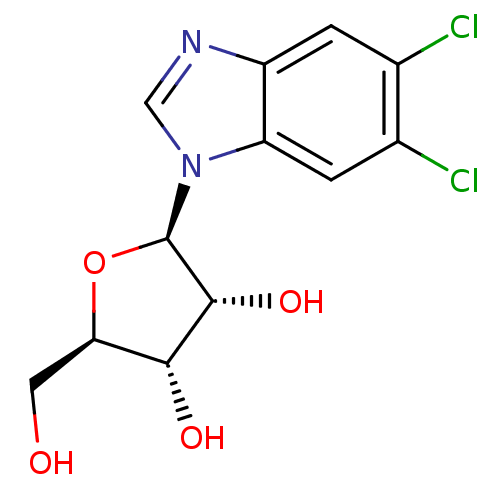 ((2R,3R,4S,5R)-2-(5,6-dichloro-1H-1,3-benzodiazol-1...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia antibodypedia antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 6.50E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Oxford Curated by ChEMBL | Assay Description Inhibition of CDK2/Cyclin A (174 to 432 amino acid residues) (unknown origin) by differential scanning fluorimetry assay | J Med Chem 56: 660-70 (2013) Article DOI: 10.1021/jm301495v BindingDB Entry DOI: 10.7270/Q28G8N18 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||