

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate receptor 2 (126/143 = 88%)† (Rattus norvegicus) | BDBM50060632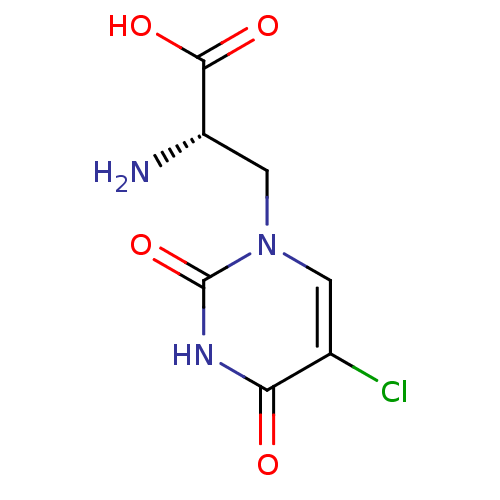 ((S)-2-Amino-3-(5-chloro-2,4-dioxo-3,4-dihydro-2H-p...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 45 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
European Research Centre for Drug Discovery and Development (NatSynDrugs) Curated by ChEMBL | Assay Description Displacement of (R,S)-[5-methyl-3H]AMPA from rat recombinant flop iGluR2(R) expressed in Sf9 cells | J Med Chem 51: 6614-8 (2008) Article DOI: 10.1021/jm800865a BindingDB Entry DOI: 10.7270/Q261117R | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor 2 (125/143 = 87%)† (Homo sapiens (Human)) | BDBM50060632 ((S)-2-Amino-3-(5-chloro-2,4-dioxo-3,4-dihydro-2H-p...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 53 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Bristol Curated by ChEMBL | Assay Description Displacement of [3H]AMPA from human Ionotropic glutamate receptor AMPA 2 expressed in HEK293 cells | J Med Chem 40: 3645-50 (1997) Article DOI: 10.1021/jm9702387 BindingDB Entry DOI: 10.7270/Q28G8MCB | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor 1 (125/143 = 87%)† (Homo sapiens (Human)) | BDBM50060632 ((S)-2-Amino-3-(5-chloro-2,4-dioxo-3,4-dihydro-2H-p...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 65 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Bristol Curated by ChEMBL | Assay Description Displacement of [3H]AMPA from human Ionotropic glutamate receptor AMPA 1 expressed in HEK293 cells | J Med Chem 40: 3645-50 (1997) Article DOI: 10.1021/jm9702387 BindingDB Entry DOI: 10.7270/Q28G8MCB | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor 4 (97/114 = 85%)† (Homo sapiens (Human)) | BDBM50060632 ((S)-2-Amino-3-(5-chloro-2,4-dioxo-3,4-dihydro-2H-p...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 451 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Bristol Curated by ChEMBL | Assay Description Displacement of [3H]AMPA from human Ionotropic glutamate receptor AMPA 4 expressed in HEK293 cells | J Med Chem 40: 3645-50 (1997) Article DOI: 10.1021/jm9702387 BindingDB Entry DOI: 10.7270/Q28G8MCB | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor 3 (138/143 = 97%)† (RAT) | BDBM50060632 ((S)-2-Amino-3-(5-chloro-2,4-dioxo-3,4-dihydro-2H-p...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 451 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
European Research Centre for Drug Discovery and Development (NatSynDrugs) Curated by ChEMBL | Assay Description Displacement of (R,S)-[5-methyl-3H]AMPA from rat recombinant flop iGluR3 expressed in Sf9 cells | J Med Chem 51: 6614-8 (2008) Article DOI: 10.1021/jm800865a BindingDB Entry DOI: 10.7270/Q261117R | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor 4 (97/114 = 85%)† (Rattus norvegicus) | BDBM50060632 ((S)-2-Amino-3-(5-chloro-2,4-dioxo-3,4-dihydro-2H-p...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 561 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
European Research Centre for Drug Discovery and Development (NatSynDrugs) Curated by ChEMBL | Assay Description Displacement of (R,S)-[5-methyl-3H]AMPA from rat recombinant flop iGluR4 expressed in Sf9 cells | J Med Chem 51: 6614-8 (2008) Article DOI: 10.1021/jm800865a BindingDB Entry DOI: 10.7270/Q261117R | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor 2 (126/143 = 88%)† (Rattus norvegicus) | BDBM50060632 ((S)-2-Amino-3-(5-chloro-2,4-dioxo-3,4-dihydro-2H-p...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | n/a | 2.11E+3 | n/a | n/a | n/a | n/a |
European Research Centre for Drug Discovery and Development (NatSynDrugs) Curated by ChEMBL | Assay Description Agonist activity at cyclothiazide-desensitized rat recombinant flip iGluR2(Q) expressed in Xenopus laevis oocytes by two electrode voltage-clamp elec... | J Med Chem 51: 6614-8 (2008) Article DOI: 10.1021/jm800865a BindingDB Entry DOI: 10.7270/Q261117R | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||