

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Histone-lysine N-methyltransferase, H3 lysine-79 specific (420/420 = 100%)† (Homo sapiens (Human)) | BDBM50431676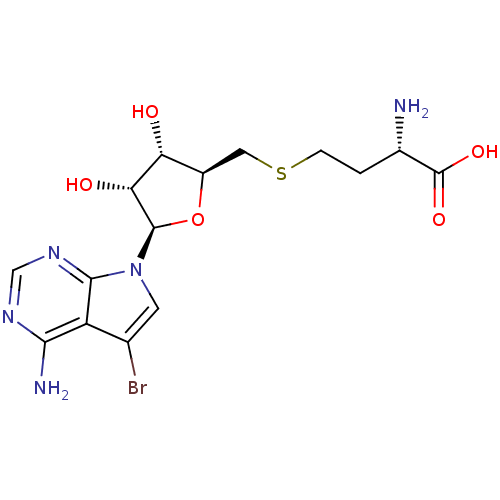 (CHEMBL2349526) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 38 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
EntreMed Inc. Curated by ChEMBL | Assay Description Competitive inhibition of human recombinant DOT1L (1 to 420 amino acid residues) overexpressed in Escherichia coli BL21 (DE3) using [3H]-SAM as subst... | Bioorg Med Chem 21: 1787-94 (2013) Article DOI: 10.1016/j.bmc.2013.01.049 BindingDB Entry DOI: 10.7270/Q24F1S41 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Histone-lysine N-methyltransferase, H3 lysine-79 specific (420/420 = 100%)† (Homo sapiens (Human)) | BDBM50431676 (CHEMBL2349526) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 77 | n/a | n/a | n/a | n/a | n/a | n/a |
Baylor College of Medicine Curated by ChEMBL | Assay Description Competitive inhibition of recombinant human DOT1L using adenosine/deazaadenosine as substrate and SAM cofactor | J Med Chem 56: 8972-83 (2013) Article DOI: 10.1021/jm4007752 BindingDB Entry DOI: 10.7270/Q2D50PDQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Histone-lysine N-methyltransferase, H3 lysine-79 specific (420/420 = 100%)† (Homo sapiens (Human)) | BDBM50431676 (CHEMBL2349526) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 77 | n/a | n/a | n/a | n/a | n/a | n/a |
EntreMed Inc. Curated by ChEMBL | Assay Description Competitive inhibition of human recombinant DOT1L (1 to 420 amino acid residues) overexpressed in Escherichia coli BL21 (DE3) using [3H]-SAM as subst... | Bioorg Med Chem 21: 1787-94 (2013) Article DOI: 10.1016/j.bmc.2013.01.049 BindingDB Entry DOI: 10.7270/Q24F1S41 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||