

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D-amino-acid oxidase (347/347 = 100%)† (Homo sapiens (Human)) | BDBM50260722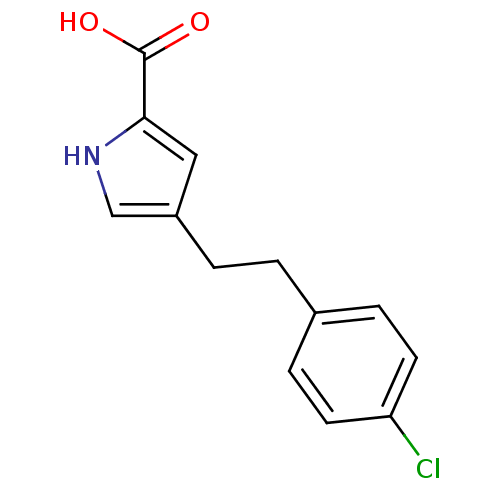 (4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | 7.20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sunovion Pharmaceuticals Inc. Curated by ChEMBL | Assay Description Competitive inhibition of human recombinant DAAO by Michaelis-Menten plot analysis in presence of D-serine | J Med Chem 56: 3710-24 (2013) Article DOI: 10.1021/jm4002583 BindingDB Entry DOI: 10.7270/Q2X92CPC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D-amino-acid oxidase (347/347 = 100%)† (Homo sapiens (Human)) | BDBM50260722 (4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | 11 | n/a | n/a | n/a | n/a | n/a | n/a |
Sunovion Pharmaceuticals Inc. Curated by ChEMBL | Assay Description Competitive inhibition of human recombinant DAAO after 1 hr by coupled enzyme assay in presence of D-serine | J Med Chem 56: 3710-24 (2013) Article DOI: 10.1021/jm4002583 BindingDB Entry DOI: 10.7270/Q2X92CPC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D-amino-acid oxidase (347/347 = 100%)† (Homo sapiens (Human)) | BDBM50260722 (4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | 238 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Sharp & Dohme Curated by ChEMBL | Assay Description Inhibition of human DAO | Bioorg Med Chem Lett 18: 3386-91 (2008) Article DOI: 10.1016/j.bmcl.2008.04.020 BindingDB Entry DOI: 10.7270/Q2WQ03KS | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D-amino-acid oxidase (347/347 = 100%)† (Homo sapiens (Human)) | BDBM50260722 (4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | n/a | 5.00E+3 | n/a | n/a | n/a | n/a | n/a |
Sunovion Pharmaceuticals Inc. Curated by ChEMBL | Assay Description Binding affinity to human recombinant DAAO at 520 nm spectral modification by stopped flow spectrophotometric analysis in presence of FAD | J Med Chem 56: 3710-24 (2013) Article DOI: 10.1021/jm4002583 BindingDB Entry DOI: 10.7270/Q2X92CPC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D-amino-acid oxidase (347/347 = 100%)† (Homo sapiens (Human)) | BDBM50260722 (4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | n/a | 2.50E+3 | n/a | n/a | n/a | n/a | n/a |
Sunovion Pharmaceuticals Inc. Curated by ChEMBL | Assay Description Binding affinity to human recombinant DAAO at 520 nm spectral modification by spectrophotometric analysis in presence of FAD | J Med Chem 56: 3710-24 (2013) Article DOI: 10.1021/jm4002583 BindingDB Entry DOI: 10.7270/Q2X92CPC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D-amino-acid oxidase (347/347 = 100%)† (Homo sapiens (Human)) | BDBM50260722 (4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | n/a | 180 | n/a | n/a | n/a | n/a | n/a |
Sunovion Pharmaceuticals Inc. Curated by ChEMBL | Assay Description Binding affinity to human recombinant DAAO at 490 nm spectral modification by spectrophotometric analysis in presence of FAD | J Med Chem 56: 3710-24 (2013) Article DOI: 10.1021/jm4002583 BindingDB Entry DOI: 10.7270/Q2X92CPC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D-amino-acid oxidase (347/347 = 100%)† (Homo sapiens (Human)) | BDBM50260722 (4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | n/a | 700 | n/a | n/a | n/a | n/a | n/a |
Sunovion Pharmaceuticals Inc. Curated by ChEMBL | Assay Description Binding affinity to human recombinant DAAO by stopped flow spectrophotometric analysis in presence of FAD | J Med Chem 56: 3710-24 (2013) Article DOI: 10.1021/jm4002583 BindingDB Entry DOI: 10.7270/Q2X92CPC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||