

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aurora kinase A (278/279 > 99%)† (Homo sapiens (Human)) | BDBM92795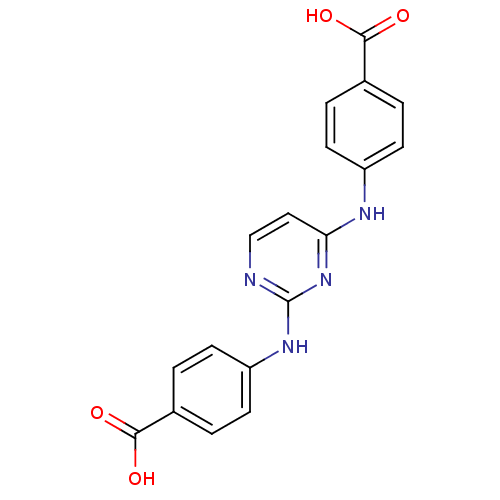 (Bisanilinopyrimidine, 3i | US9249124, 10) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase MMDB PC cid PC sid PDB UniChem Similars | PDB US Patent | n/a | n/a | 256 | n/a | n/a | n/a | n/a | 7.4 | 25 |
H. Lee Moffitt Cancer Center and Research Institute, Inc. US Patent | Assay Description Reactions were carried out at room temperature in 15 mM HEPES buffer (pH 7.4) containing 20 mM NaCl, 1 mM EGTA, 0.02% Tween 20, 10 mM MgCl2, 5% (v/v)... | US Patent US9249124 (2016) BindingDB Entry DOI: 10.7270/Q2J1020R | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Aurora kinase A (278/279 > 99%)† (Homo sapiens (Human)) | BDBM92795 (Bisanilinopyrimidine, 3i | US9249124, 10) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 256 | n/a | n/a | n/a | n/a | n/a | n/a |
Moffitt Cancer Center | Assay Description In vitro enzyme activity assay using Aurora Kinase A. | J Med Chem 55: 7392-416 (2012) Article DOI: 10.1021/jm300334d BindingDB Entry DOI: 10.7270/Q2V986NH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||