

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bromodomain-containing protein 4 (112/112 = 100%)† (Homo sapiens (Human)) | BDBM50092310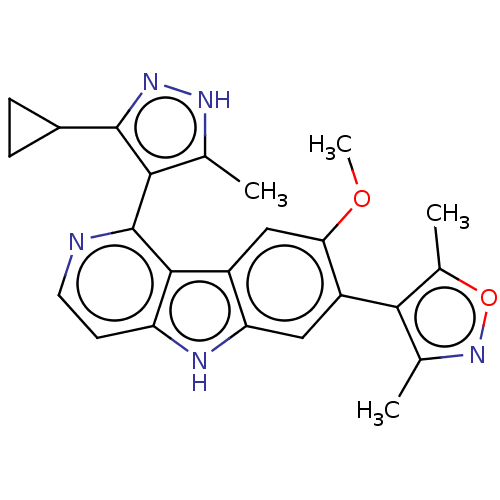 (CHEMBL3581661 | US9675697, Cpd. No. 23) | PDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 12 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Michigan Curated by ChEMBL | Assay Description Displacement of FAM-labeled ZBA248 from BRD4 BD2 (333 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by f... | J Med Chem 58: 4927-39 (2015) Article DOI: 10.1021/acs.jmedchem.5b00613 BindingDB Entry DOI: 10.7270/Q2XD13FK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Bromodomain-containing protein 4 (112/112 = 100%)† (Homo sapiens (Human)) | BDBM50092310 (CHEMBL3581661 | US9675697, Cpd. No. 23) | PDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 25 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Michigan Curated by ChEMBL | Assay Description Displacement of FAM-labeled ZBA248 from BRD4 BD1 (44 to 168 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fl... | J Med Chem 58: 4927-39 (2015) Article DOI: 10.1021/acs.jmedchem.5b00613 BindingDB Entry DOI: 10.7270/Q2XD13FK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Bromodomain-containing protein 4 (112/112 = 100%)† (Homo sapiens (Human)) | BDBM50092310 (CHEMBL3581661 | US9675697, Cpd. No. 23) | PDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 37 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Michigan Curated by ChEMBL | Assay Description Displacement of FAM-labeled ZBA248 from BRD4 BD2 (333 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by f... | J Med Chem 58: 4927-39 (2015) Article DOI: 10.1021/acs.jmedchem.5b00613 BindingDB Entry DOI: 10.7270/Q2XD13FK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Bromodomain-containing protein 4 (112/112 = 100%)† (Homo sapiens (Human)) | BDBM50092310 (CHEMBL3581661 | US9675697, Cpd. No. 23) | PDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 76 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Michigan Curated by ChEMBL | Assay Description Displacement of FAM-labeled ZBA248 from BRD4 BD1 (44 to 168 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fl... | J Med Chem 58: 4927-39 (2015) Article DOI: 10.1021/acs.jmedchem.5b00613 BindingDB Entry DOI: 10.7270/Q2XD13FK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Bromodomain-containing protein 4 (112/112 = 100%)† (Homo sapiens (Human)) | BDBM50092310 (CHEMBL3581661 | US9675697, Cpd. No. 23) | PDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 47 | n/a | n/a | n/a | n/a | n/a |
University of Michigan Curated by ChEMBL | Assay Description Binding affinity to biotinylated BRD4 BD1 (44 to 168 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometr... | J Med Chem 58: 4927-39 (2015) Article DOI: 10.1021/acs.jmedchem.5b00613 BindingDB Entry DOI: 10.7270/Q2XD13FK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Bromodomain-containing protein 4 (112/112 = 100%)† (Homo sapiens (Human)) | BDBM50092310 (CHEMBL3581661 | US9675697, Cpd. No. 23) | PDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 45 | n/a | n/a | n/a | n/a | n/a |
University of Michigan Curated by ChEMBL | Assay Description Binding affinity to biotinylated BRD4 BD2 (333 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferomet... | J Med Chem 58: 4927-39 (2015) Article DOI: 10.1021/acs.jmedchem.5b00613 BindingDB Entry DOI: 10.7270/Q2XD13FK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||