

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adenosylhomocysteine nucleosidase (216/229 = 94%)† (Helicobacter pylori) | BDBM36498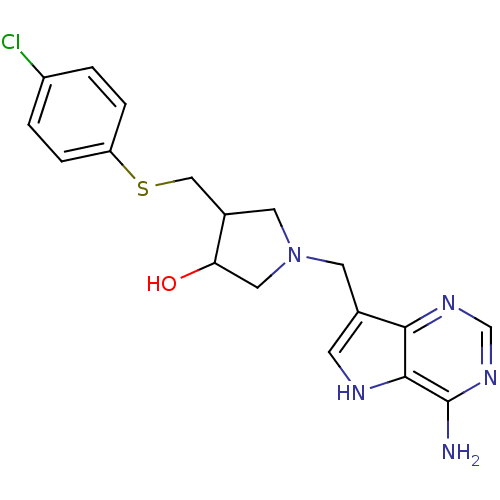 (DADMe-ImmA-p-ClPh) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 0.570 | n/a | n/a | n/a | 7.5 | 25 |
Albert Einstein College of Medicine | Assay Description Enzyme activity assay using various bacterial species of 5'-Methylthioadenosine nucleosidases (MTANs) excepted for MTAP, for human MTAP. | ACS Chem Biol 2: 725-34 (2007) Article DOI: 10.1021/cb700166z BindingDB Entry DOI: 10.7270/Q2QF8R7W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||