 Found 6 hits Enzyme Inhibition Constant Data
Found 6 hits Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Serine/threonine-protein kinase mTOR
(92/93 = 99%)†
(Homo sapiens (Human)) | BDBM36609
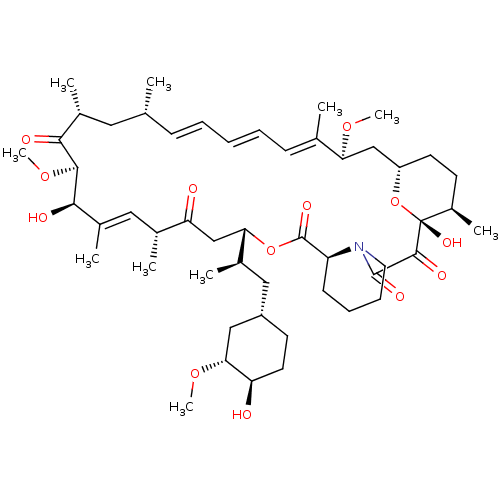
(Rapamycin C-7, analog 4 | SIROLIMUS | US11603377, ...)Show SMILES CO[C@@H]1C[C@H](C[C@@H](C)[C@@H]2CC(=O)[C@H](C)\C=C(C)\[C@@H](O)[C@@H](OC)C(=O)[C@H](C)C[C@H](C)\C=C\C=C\C=C(C)\[C@H](C[C@@H]3CC[C@@H](C)[C@@](O)(O3)C(=O)C(=O)N3CCCC[C@H]3C(=O)O2)OC)CC[C@H]1O |c:14,33,t:29,31| Show InChI InChI=1S/C51H79NO13/c1-30-16-12-11-13-17-31(2)42(61-8)28-38-21-19-36(7)51(60,65-38)48(57)49(58)52-23-15-14-18-39(52)50(59)64-43(33(4)26-37-20-22-40(53)44(27-37)62-9)29-41(54)32(3)25-35(6)46(56)47(63-10)45(55)34(5)24-30/h11-13,16-17,25,30,32-34,36-40,42-44,46-47,53,56,60H,14-15,18-24,26-29H2,1-10H3/b13-11+,16-12+,31-17+,35-25+/t30-,32-,33-,34-,36-,37+,38+,39+,40-,42+,43+,44-,46-,47+,51-/m1/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
KEGG
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 0.100 | n/a | n/a | n/a | n/a | n/a | n/a |
Stanford University
Curated by ChEMBL
| Assay Description
Inhibition of mTOR kinase expressed in human HEK293 cells by western blot analysis |
J Biol Chem 282: 13395-401 (2007)
Article DOI: 10.1074/jbc.M700498200
BindingDB Entry DOI: 10.7270/Q2J1042G |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Serine/threonine-protein kinase mTOR
(92/93 = 99%)†
(Homo sapiens (Human)) | BDBM36609

(Rapamycin C-7, analog 4 | SIROLIMUS | US11603377, ...)Show SMILES CO[C@@H]1C[C@H](C[C@@H](C)[C@@H]2CC(=O)[C@H](C)\C=C(C)\[C@@H](O)[C@@H](OC)C(=O)[C@H](C)C[C@H](C)\C=C\C=C\C=C(C)\[C@H](C[C@@H]3CC[C@@H](C)[C@@](O)(O3)C(=O)C(=O)N3CCCC[C@H]3C(=O)O2)OC)CC[C@H]1O |c:14,33,t:29,31| Show InChI InChI=1S/C51H79NO13/c1-30-16-12-11-13-17-31(2)42(61-8)28-38-21-19-36(7)51(60,65-38)48(57)49(58)52-23-15-14-18-39(52)50(59)64-43(33(4)26-37-20-22-40(53)44(27-37)62-9)29-41(54)32(3)25-35(6)46(56)47(63-10)45(55)34(5)24-30/h11-13,16-17,25,30,32-34,36-40,42-44,46-47,53,56,60H,14-15,18-24,26-29H2,1-10H3/b13-11+,16-12+,31-17+,35-25+/t30-,32-,33-,34-,36-,37+,38+,39+,40-,42+,43+,44-,46-,47+,51-/m1/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
KEGG
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 0.450 | n/a | n/a | n/a | n/a | n/a | n/a |
The Scripps Research Institute
Curated by ChEMBL
| Assay Description
Inhibitory concentration against FKBP12 receptor |
J Med Chem 48: 5613-38 (2005)
Article DOI: 10.1021/jm050524f
BindingDB Entry DOI: 10.7270/Q2V988V1 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Serine/threonine-protein kinase mTOR
(92/93 = 99%)†
(Homo sapiens (Human)) | BDBM36609

(Rapamycin C-7, analog 4 | SIROLIMUS | US11603377, ...)Show SMILES CO[C@@H]1C[C@H](C[C@@H](C)[C@@H]2CC(=O)[C@H](C)\C=C(C)\[C@@H](O)[C@@H](OC)C(=O)[C@H](C)C[C@H](C)\C=C\C=C\C=C(C)\[C@H](C[C@@H]3CC[C@@H](C)[C@@](O)(O3)C(=O)C(=O)N3CCCC[C@H]3C(=O)O2)OC)CC[C@H]1O |c:14,33,t:29,31| Show InChI InChI=1S/C51H79NO13/c1-30-16-12-11-13-17-31(2)42(61-8)28-38-21-19-36(7)51(60,65-38)48(57)49(58)52-23-15-14-18-39(52)50(59)64-43(33(4)26-37-20-22-40(53)44(27-37)62-9)29-41(54)32(3)25-35(6)46(56)47(63-10)45(55)34(5)24-30/h11-13,16-17,25,30,32-34,36-40,42-44,46-47,53,56,60H,14-15,18-24,26-29H2,1-10H3/b13-11+,16-12+,31-17+,35-25+/t30-,32-,33-,34-,36-,37+,38+,39+,40-,42+,43+,44-,46-,47+,51-/m1/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
KEGG
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
PubMed
| n/a | n/a | 3.5 | n/a | n/a | n/a | n/a | n/a | n/a |
Glaxo Wellcome
Curated by ChEMBL
| Assay Description
The inhibitory activity by using FK506 binding protein 12 SPA binding assay |
Bioorg Med Chem Lett 10: 1007-10 (2000)
BindingDB Entry DOI: 10.7270/Q28K7BCP |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Serine/threonine-protein kinase mTOR
(92/93 = 99%)†
(Homo sapiens (Human)) | BDBM36609

(Rapamycin C-7, analog 4 | SIROLIMUS | US11603377, ...)Show SMILES CO[C@@H]1C[C@H](C[C@@H](C)[C@@H]2CC(=O)[C@H](C)\C=C(C)\[C@@H](O)[C@@H](OC)C(=O)[C@H](C)C[C@H](C)\C=C\C=C\C=C(C)\[C@H](C[C@@H]3CC[C@@H](C)[C@@](O)(O3)C(=O)C(=O)N3CCCC[C@H]3C(=O)O2)OC)CC[C@H]1O |c:14,33,t:29,31| Show InChI InChI=1S/C51H79NO13/c1-30-16-12-11-13-17-31(2)42(61-8)28-38-21-19-36(7)51(60,65-38)48(57)49(58)52-23-15-14-18-39(52)50(59)64-43(33(4)26-37-20-22-40(53)44(27-37)62-9)29-41(54)32(3)25-35(6)46(56)47(63-10)45(55)34(5)24-30/h11-13,16-17,25,30,32-34,36-40,42-44,46-47,53,56,60H,14-15,18-24,26-29H2,1-10H3/b13-11+,16-12+,31-17+,35-25+/t30-,32-,33-,34-,36-,37+,38+,39+,40-,42+,43+,44-,46-,47+,51-/m1/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
KEGG
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 430 | n/a | n/a | n/a | n/a | n/a | n/a |
Mansoura University
Curated by ChEMBL
| Assay Description
Inhibition of mTOR (unknown origin) incubated for 1 hr by ELISA |
Eur J Med Chem 156: 304-315 (2018)
Article DOI: 10.1016/j.ejmech.2018.07.004
BindingDB Entry DOI: 10.7270/Q2J9692K |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Serine/threonine-protein kinase mTOR
(92/93 = 99%)†
(Homo sapiens (Human)) | BDBM36609

(Rapamycin C-7, analog 4 | SIROLIMUS | US11603377, ...)Show SMILES CO[C@@H]1C[C@H](C[C@@H](C)[C@@H]2CC(=O)[C@H](C)\C=C(C)\[C@@H](O)[C@@H](OC)C(=O)[C@H](C)C[C@H](C)\C=C\C=C\C=C(C)\[C@H](C[C@@H]3CC[C@@H](C)[C@@](O)(O3)C(=O)C(=O)N3CCCC[C@H]3C(=O)O2)OC)CC[C@H]1O |c:14,33,t:29,31| Show InChI InChI=1S/C51H79NO13/c1-30-16-12-11-13-17-31(2)42(61-8)28-38-21-19-36(7)51(60,65-38)48(57)49(58)52-23-15-14-18-39(52)50(59)64-43(33(4)26-37-20-22-40(53)44(27-37)62-9)29-41(54)32(3)25-35(6)46(56)47(63-10)45(55)34(5)24-30/h11-13,16-17,25,30,32-34,36-40,42-44,46-47,53,56,60H,14-15,18-24,26-29H2,1-10H3/b13-11+,16-12+,31-17+,35-25+/t30-,32-,33-,34-,36-,37+,38+,39+,40-,42+,43+,44-,46-,47+,51-/m1/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
KEGG
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Celgene Corporation
Curated by ChEMBL
| Assay Description
Inhibition of mTOR-mediated Akt phosphorylation in human PC3 cells |
Bioorg Med Chem Lett 21: 6793-9 (2011)
Article DOI: 10.1016/j.bmcl.2011.09.035
BindingDB Entry DOI: 10.7270/Q28W3DR1 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Serine/threonine-protein kinase mTOR
(92/93 = 99%)†
(Homo sapiens (Human)) | BDBM36609

(Rapamycin C-7, analog 4 | SIROLIMUS | US11603377, ...)Show SMILES CO[C@@H]1C[C@H](C[C@@H](C)[C@@H]2CC(=O)[C@H](C)\C=C(C)\[C@@H](O)[C@@H](OC)C(=O)[C@H](C)C[C@H](C)\C=C\C=C\C=C(C)\[C@H](C[C@@H]3CC[C@@H](C)[C@@](O)(O3)C(=O)C(=O)N3CCCC[C@H]3C(=O)O2)OC)CC[C@H]1O |c:14,33,t:29,31| Show InChI InChI=1S/C51H79NO13/c1-30-16-12-11-13-17-31(2)42(61-8)28-38-21-19-36(7)51(60,65-38)48(57)49(58)52-23-15-14-18-39(52)50(59)64-43(33(4)26-37-20-22-40(53)44(27-37)62-9)29-41(54)32(3)25-35(6)46(56)47(63-10)45(55)34(5)24-30/h11-13,16-17,25,30,32-34,36-40,42-44,46-47,53,56,60H,14-15,18-24,26-29H2,1-10H3/b13-11+,16-12+,31-17+,35-25+/t30-,32-,33-,34-,36-,37+,38+,39+,40-,42+,43+,44-,46-,47+,51-/m1/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
KEGG
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 0.600 | n/a | n/a | n/a | n/a | n/a |
University of Basel
Curated by ChEMBL
| Assay Description
Binding affinity to mTOR FKBP12 site (unknown origin) |
J Med Chem 61: 10084-10105 (2018)
Article DOI: 10.1021/acs.jmedchem.8b01262
BindingDB Entry DOI: 10.7270/Q2X92F07 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data