

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280553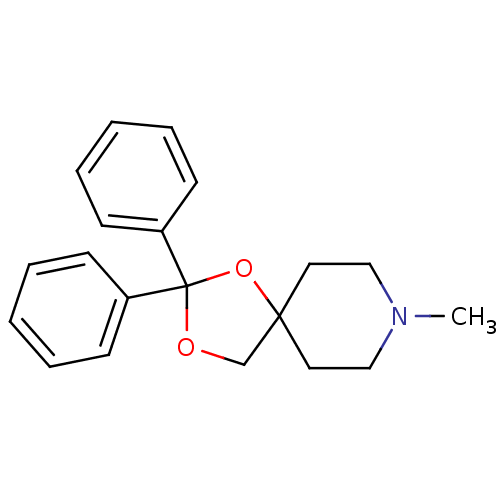 (8-Methyl-2,2-diphenyl-1,3-dioxa-8-aza-spiro[4.5]de...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 0.600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM73209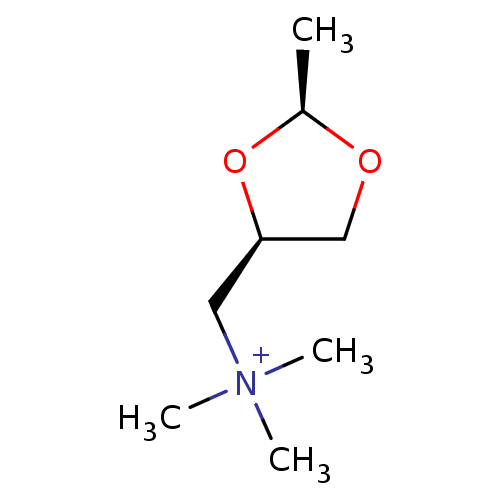 ((+)-CIS-DIOXOLANE | CHEMBL105457 | MLS000069362 | ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article | 6.40 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280553 (8-Methyl-2,2-diphenyl-1,3-dioxa-8-aza-spiro[4.5]de...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 8.10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50006243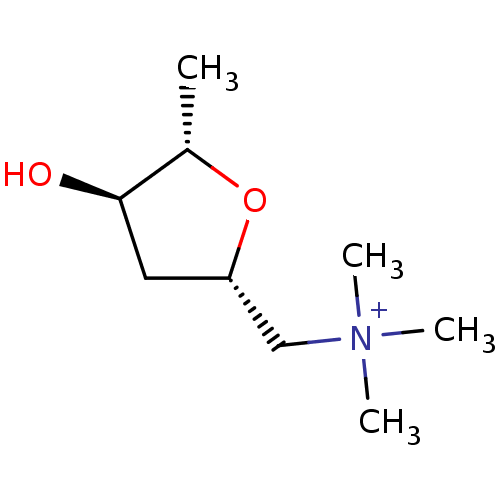 (((2S,4R,5S)-4-Hydroxy-5-methyl-tetrahydro-furan-2-...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG PC cid PC sid UniChem Similars | Article | 12 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280540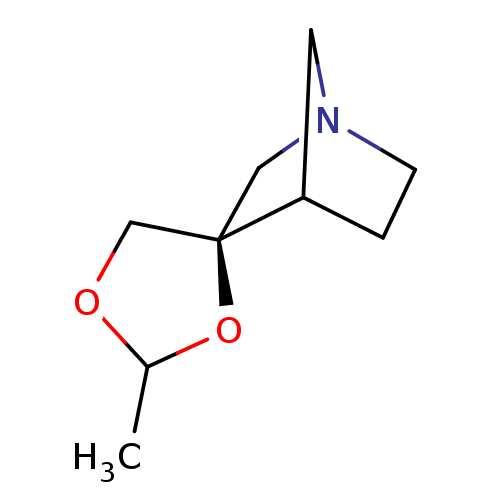 (Azaspirodioxolane analogue | CHEMBL170388) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 30 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280536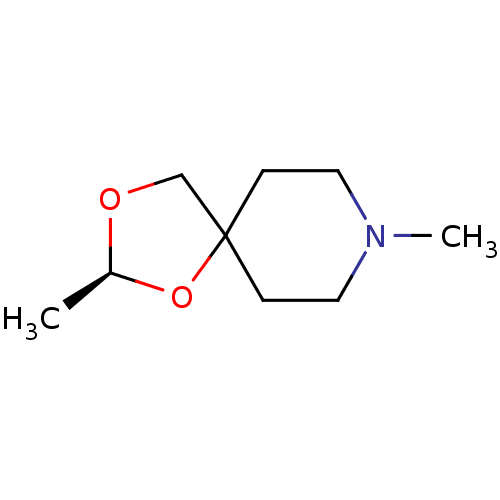 ((S)-2,8-Dimethyl-1,3-dioxa-8-aza-spiro[4.5]decane ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 85 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280558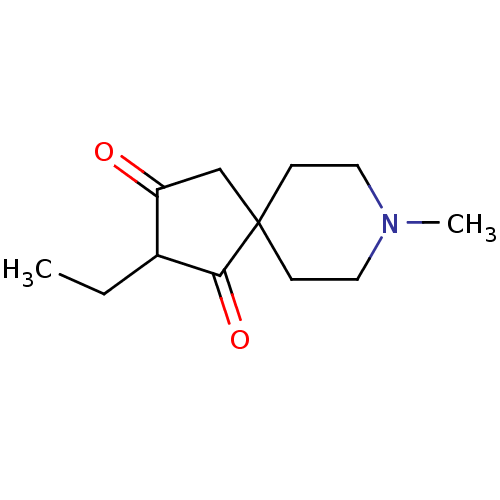 (2-Ethyl-8-methyl-8-aza-spiro[4.5]decane-1,3-dione ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 87 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50005848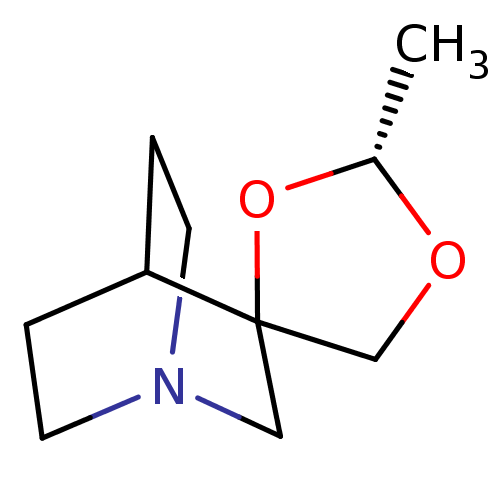 ((2'R)-2'-methylspiro[4-azabicyclo[2.2.2]octane-2,4...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 90 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50034640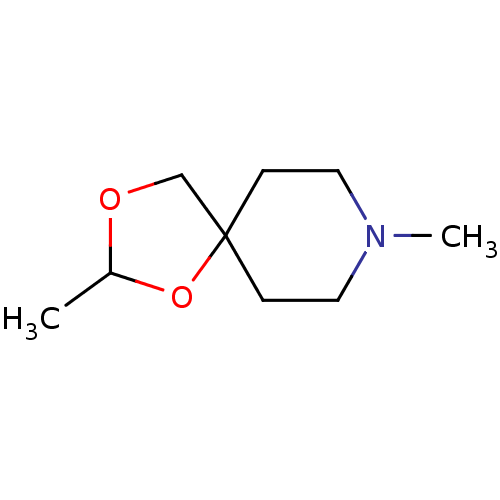 (2,8-Dimethyl-1,3-dioxa-8-aza-spiro[4.5]decane | CH...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449645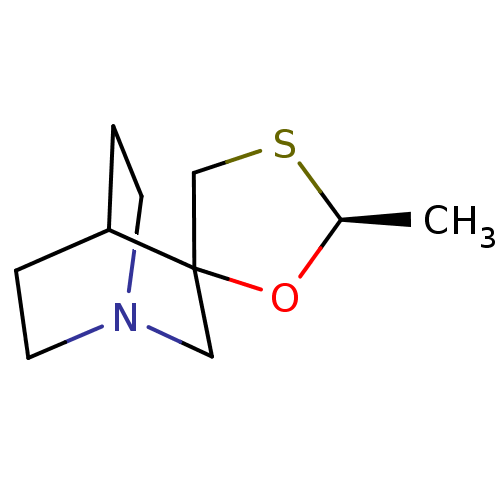 (AF-102B | CHEMBL2021361 | FKS-508 | SND-5008 | SNI...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article | 105 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280545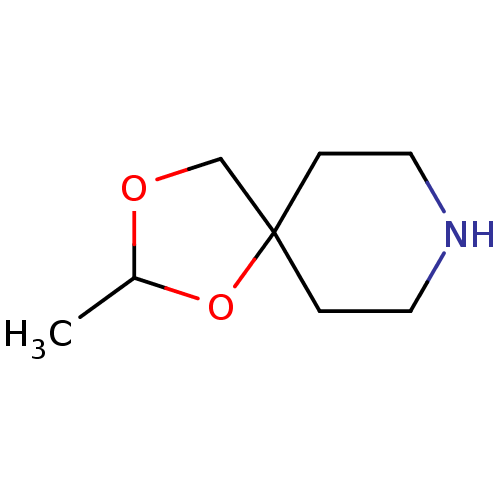 (2-Methyl-1,3-dioxa-8-aza-spiro[4.5]decane | CHEMBL...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article | 145 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280556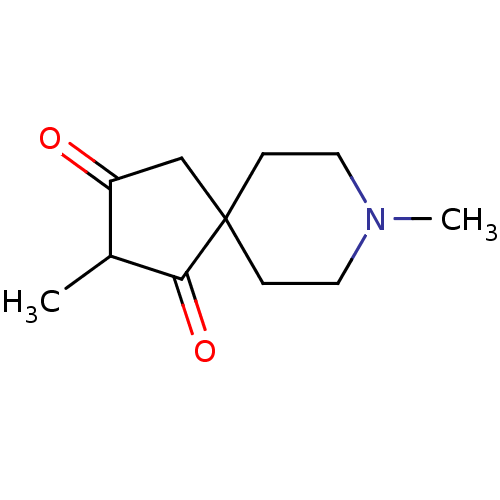 (2,8-Dimethyl-8-aza-spiro[4.5]decane-1,3-dione | CH...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 175 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280550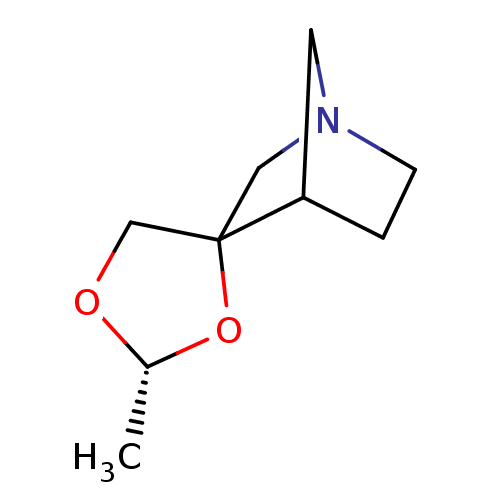 (Azaspirodioxolane analogue | CHEMBL170489) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 180 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280539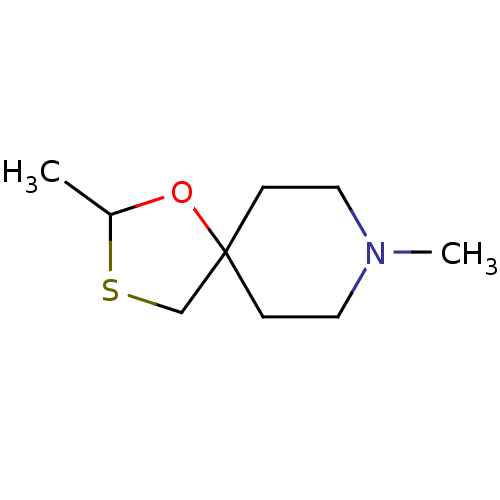 (2,8-Dimethyl-1-oxa-3-thia-8-aza-spiro[4.5]decane |...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 185 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested in vitro for its muscarinic m1 receptor agonist potency in the ileum model as a functional measure of peripheral muscarinic m1 re... | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449644 (CHEMBL2371594) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280555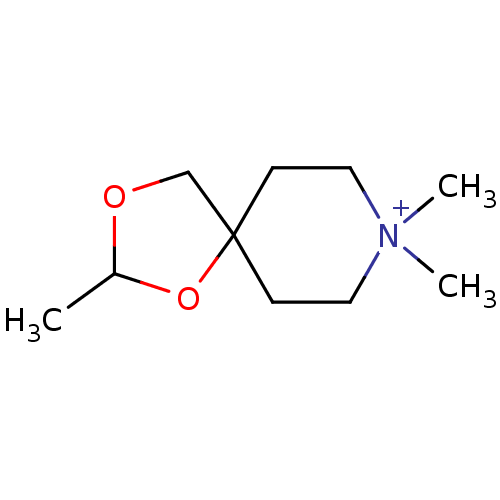 (2,8,8-Trimethyl-1,3-dioxa-8-azonia-spiro[4.5]decan...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 210 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280547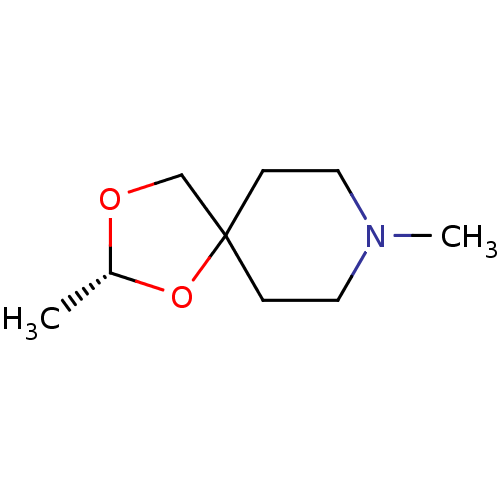 ((R)-2,8-Dimethyl-1,3-dioxa-8-aza-spiro[4.5]decane ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 240 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280554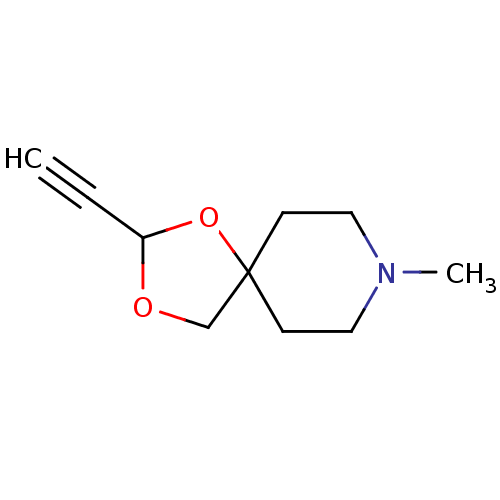 (2-Ethynyl-8-methyl-1,3-dioxa-8-aza-spiro[4.5]decan...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 295 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449645 (AF-102B | CHEMBL2021361 | FKS-508 | SND-5008 | SNI...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article | 430 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280535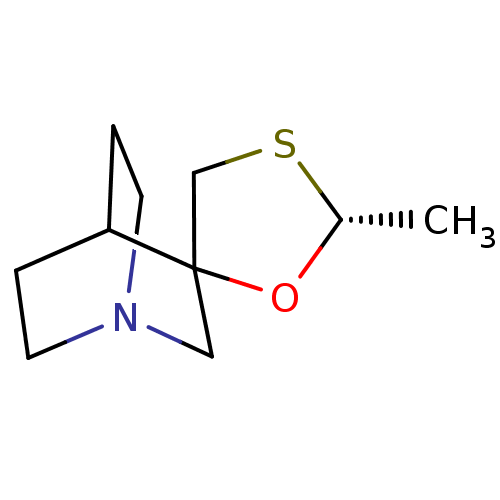 ((2'R)-2'-methylspiro[4-azabicyclo[2.2.2]octane-2,5...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article | 515 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280552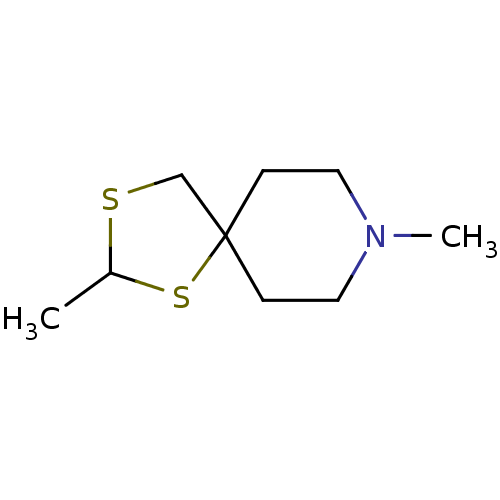 (2,8-Dimethyl-1,3-dithia-8-aza-spiro[4.5]decane | C...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 530 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449646 (CHEMBL2021360) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 585 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280546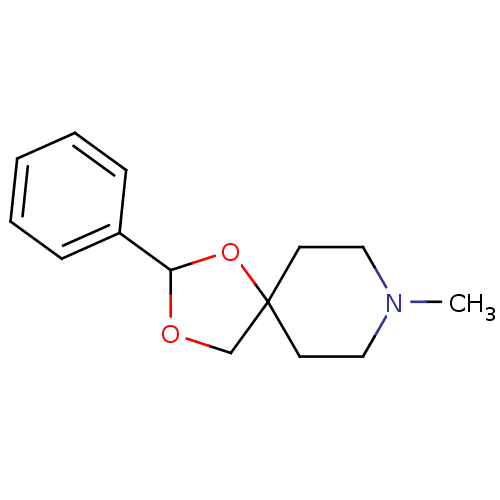 (8-Methyl-2-phenyl-1,3-dioxa-8-aza-spiro[4.5]decane...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 620 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280542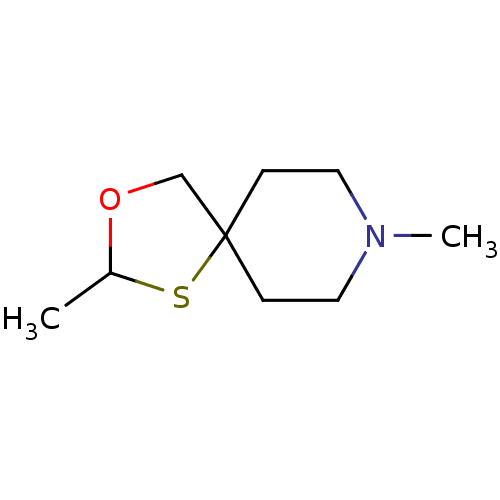 (2,8-Dimethyl-3-oxa-1-thia-8-aza-spiro[4.5]decane |...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 720 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280537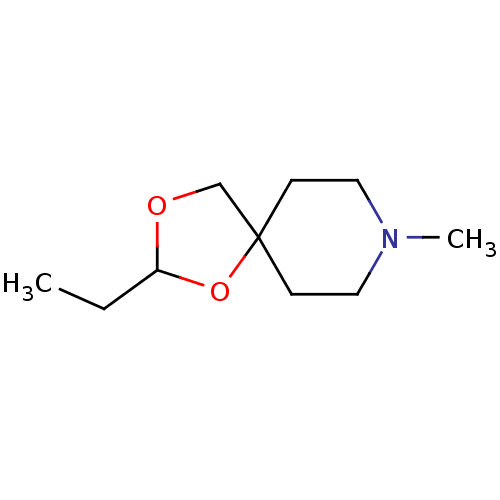 (2-Ethyl-8-methyl-1,3-dioxa-8-aza-spiro[4.5]decane ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 745 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280558 (2-Ethyl-8-methyl-8-aza-spiro[4.5]decane-1,3-dione ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 765 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280548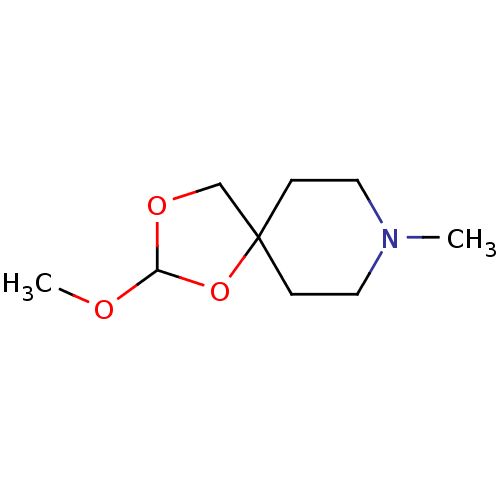 (2-Methoxy-8-methyl-1,3-dioxa-8-aza-spiro[4.5]decan...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 870 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449644 (CHEMBL2371594) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 960 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280557 (8-Methyl-1,3-dioxa-8-aza-spiro[4.5]decane | CHEMBL...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 1.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280535 ((2'R)-2'-methylspiro[4-azabicyclo[2.2.2]octane-2,5...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article | 1.25E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280537 (2-Ethyl-8-methyl-1,3-dioxa-8-aza-spiro[4.5]decane ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 1.33E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50005848 ((2'R)-2'-methylspiro[4-azabicyclo[2.2.2]octane-2,4...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 1.58E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280549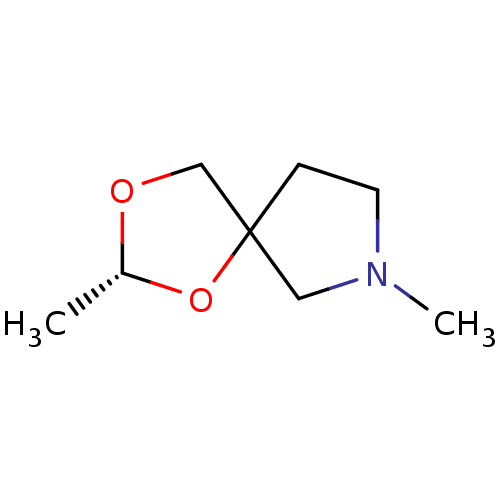 ((Trans)-(R)-2,7-Dimethyl-1,3-dioxa-7-aza-spiro[4.4...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 1.75E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280549 ((Trans)-(R)-2,7-Dimethyl-1,3-dioxa-7-aza-spiro[4.4...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 1.75E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50067479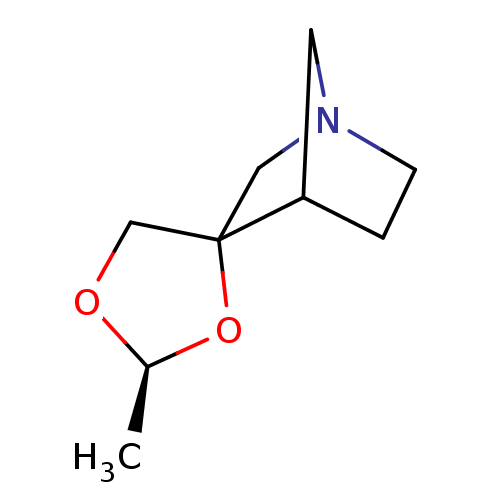 (2'-methyl-(2'S)-spiro[4-azabicyclo[2.2.1]heptane-2...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 1.85E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280540 (Azaspirodioxolane analogue | CHEMBL170388) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 1.95E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM73209 ((+)-CIS-DIOXOLANE | CHEMBL105457 | MLS000069362 | ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article | 2.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280552 (2,8-Dimethyl-1,3-dithia-8-aza-spiro[4.5]decane | C...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 2.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280539 (2,8-Dimethyl-1-oxa-3-thia-8-aza-spiro[4.5]decane |...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 2.65E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280546 (8-Methyl-2-phenyl-1,3-dioxa-8-aza-spiro[4.5]decane...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 2.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280554 (2-Ethynyl-8-methyl-1,3-dioxa-8-aza-spiro[4.5]decan...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 3.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280542 (2,8-Dimethyl-3-oxa-1-thia-8-aza-spiro[4.5]decane |...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 3.16E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested in vitro for its muscarinic m1 receptor agonist potency in the ileum model as a functional measure of peripheral muscarinic m1 re... | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280556 (2,8-Dimethyl-8-aza-spiro[4.5]decane-1,3-dione | CH...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 4.05E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280555 (2,8,8-Trimethyl-1,3-dioxa-8-azonia-spiro[4.5]decan...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 5.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280536 ((S)-2,8-Dimethyl-1,3-dioxa-8-aza-spiro[4.5]decane ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 5.33E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50006243 (((2S,4R,5S)-4-Hydroxy-5-methyl-tetrahydro-furan-2-...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG PC cid PC sid UniChem Similars | Article | 5.68E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50034640 (2,8-Dimethyl-1,3-dioxa-8-aza-spiro[4.5]decane | CH...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 5.83E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280550 (Azaspirodioxolane analogue | CHEMBL170489) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 6.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449646 (CHEMBL2021360) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 6.70E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280547 ((R)-2,8-Dimethyl-1,3-dioxa-8-aza-spiro[4.5]decane ...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 7.69E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449647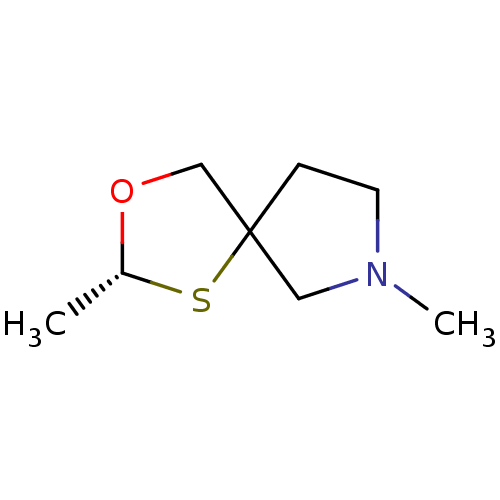 (CHEMBL2021359) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | 9.15E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280557 (8-Methyl-1,3-dioxa-8-aza-spiro[4.5]decane | CHEMBL...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50067479 (2'-methyl-(2'S)-spiro[4-azabicyclo[2.2.1]heptane-2...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280538 (2,2,8-Trimethyl-1,3-dioxa-8-aza-spiro[4.5]decane |...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280538 (2,2,8-Trimethyl-1,3-dioxa-8-aza-spiro[4.5]decane |...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested in vitro for its efficacy in the ileum model as a functional measure of peripheral muscarinic m1 receptor activity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50449647 (CHEMBL2021359) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280548 (2-Methoxy-8-methyl-1,3-dioxa-8-aza-spiro[4.5]decan...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280545 (2-Methyl-1,3-dioxa-8-aza-spiro[4.5]decane | CHEMBL...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50280549 ((Trans)-(R)-2,7-Dimethyl-1,3-dioxa-7-aza-spiro[4.4...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro 100 nM cortex affinity to displace [3H]- -pirenzepine providing a measure of muscarinic m1 receptor antagonist affinity | Bioorg Med Chem Lett 2: 815-820 (1992) Article DOI: 10.1016/S0960-894X(00)80537-5 BindingDB Entry DOI: 10.7270/Q23X8734 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||