

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50001884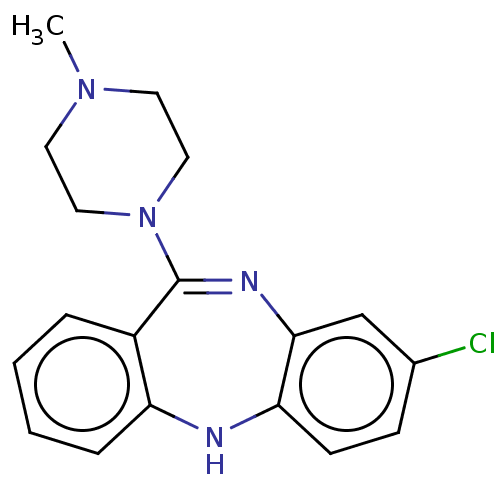 (2-[4-(4-Methyl-benzyl)-piperazin-1-yl]-1-(2-methyl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | 12 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50028979 (8-Chloro-5-methyl-11-(4-methyl-piperazin-1-yl)-5H-...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | 12 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50028601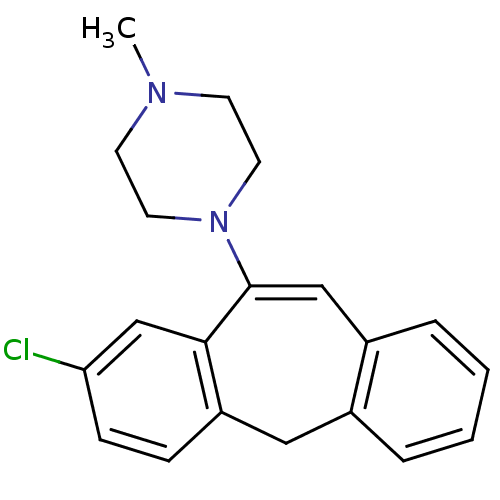 (1-(8-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-4-me...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 39 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50028980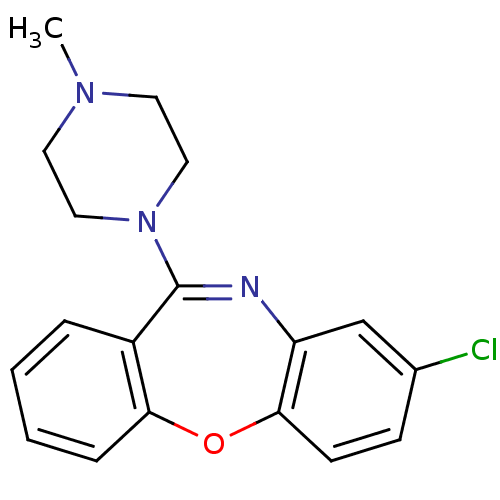 (8-Chloro-11-(4-methyl-piperazin-1-yl)-dibenzo[b,f]...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | 49 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50028601 (1-(8-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-4-me...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 290 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50028599 (1-(2-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-4-me...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 342 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50223642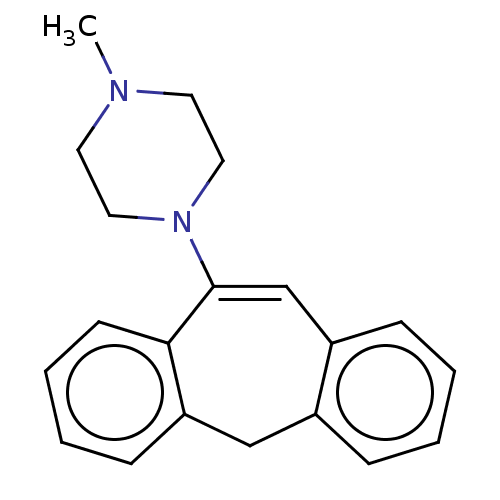 (CHEMBL7821) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 610 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50028976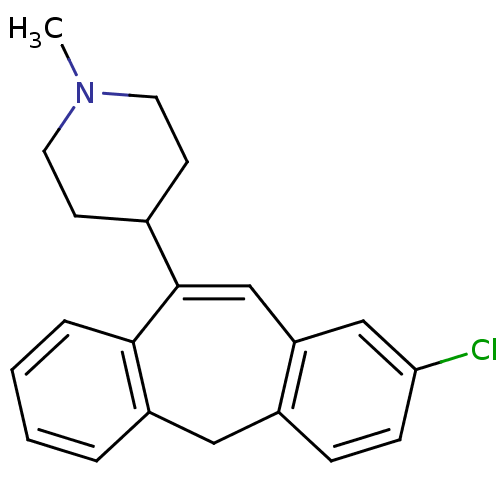 (4-(2-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-1-me...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | PubMed | n/a | n/a | 622 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50028599 (1-(2-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-4-me...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 670 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50028976 (4-(2-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-1-me...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | PubMed | n/a | n/a | 678 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50010594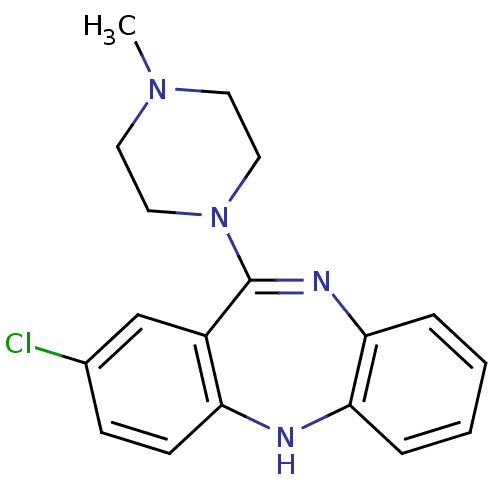 (2-Chloro-11-(4-methyl-piperazin-1-yl)-5H-dibenzo[b...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | 758 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50223642 (CHEMBL7821) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 808 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50028980 (8-Chloro-11-(4-methyl-piperazin-1-yl)-dibenzo[b,f]...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | 2.48E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50001884 (2-[4-(4-Methyl-benzyl)-piperazin-1-yl]-1-(2-methyl...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | 4.31E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50028978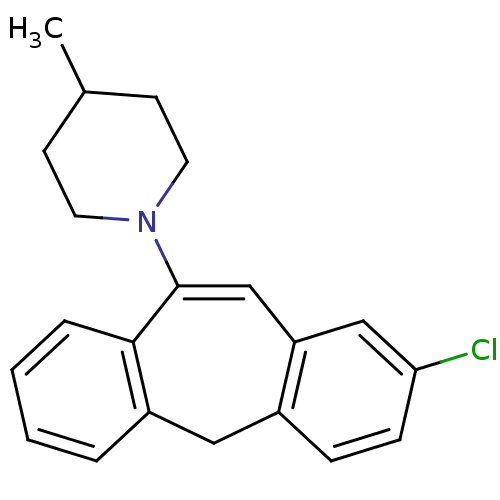 (1-(2-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-4-me...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50028978 (1-(2-Chloro-5H-dibenzo[a,d]cyclohepten-10-yl)-4-me...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 (RAT) | BDBM50223641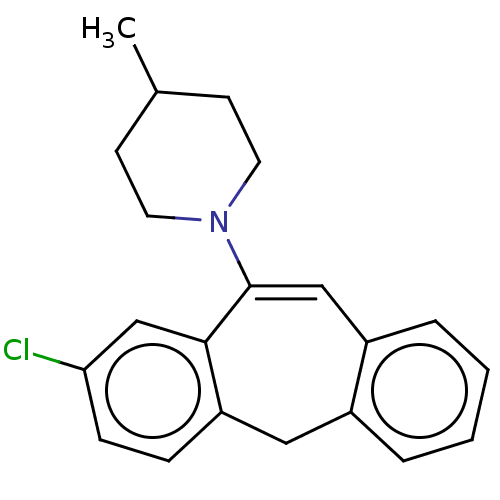 (CHEMBL266816) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Concentration required (in vitro) to displace 50% specific binding of [3H]clozapine to Muscarinic acetylcholine receptor in rat brain | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor (Rattus norvegicus (rat)-RAT-Rattus norvegicus (Rat...) | BDBM50028979 (8-Chloro-5-methyl-11-(4-methyl-piperazin-1-yl)-5H-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | 3.24E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Relative affinity for dopamine receptor by displacement of [3H]spiroperidol (2.2 nM) from (in vitro) dopamine binding sites in rat caudate nuclei | J Med Chem 24: 1021-6 (1981) BindingDB Entry DOI: 10.7270/Q2348NKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||