

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Major Urinary Protein (MUP-1) | ||
| Syringe Reactant: | BDBM12030 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 11/12/06 | ||
| ΔG°: | -8.11±0.0700 (kcal/mole) | ||
| pH: | 7.4±n/a | ||
| Log10Kb: | 5.74± n/a | ||
| Temperature: | 308±n/a (K) | ||
| ΔH° : | n/a | ||
| ΔHobs : | -10.7±0.100 (kcal/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | 0.97 | ||
| ΔS° : | -0.0100±0 (kcal/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Bingham, RJ; Findlay, JB; Hsieh, SY; Kalverda, AP; Kjellberg, A; Perazzolo, C; Phillips, SE; Seshadri, K; Trinh, CH; Turnbull, WB; Bodenhausen, G; Homans, SW Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein J Am Chem Soc126:1675-1681 (2004) Article Bingham, RJ; Findlay, JB; Hsieh, SY; Kalverda, AP; Kjellberg, A; Perazzolo, C; Phillips, SE; Seshadri, K; Trinh, CH; Turnbull, WB; Bodenhausen, G; Homans, SW Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein J Am Chem Soc126:1675-1681 (2004) Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| Major Urinary Protein (MUP-1) | |||
| Source: | The MUP-1 gene was cloned and expressed in E. coli cells. | ||
| Purity: | n/a | ||
| Prep. Method: | MUP-I was purified using Ni-NTA resin. To remove endogenous bound ligands, an ethanol precipitation step was utilized involving addition of 2 volumes of absolute ethanol per volume of protein solution, followed by incubation at 4 °C for 2 h. After centrifugation at 5000g, the pellet was lyophilised, resuspended in PBS pH 7.4, and dialyzed overnight against the same buffer. | ||
| Name: | Major Urinary Protein (MUP-1) | ||
| Synonyms: | MUP1_MOUSE | Major urinary protein 1 precursor | Mup1 | ||
| Type: | Other Protein Type | ||
| Mol. Mass.: | 20640.17 | ||
| Organism: | Mus musculus (mouse) | ||
| Description: | n/a | ||
| Residue: | 180 | ||
| Sequence: |
| ||
| BDBM12030 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM12030 | ||
| Synonyms: | 2-ISOPROPYL-3-METHOXYPYRAZINE | 2-methoxy-3-(propan-2-yl)pyrazine | IPMP | ||
| Type | Small organic molecule | ||
| Emp. Form. | C8H12N2O | ||
| Mol. Mass. | 152.1937 | ||
| SMILES | COc1nccnc1C(C)C | ||
| Structure | 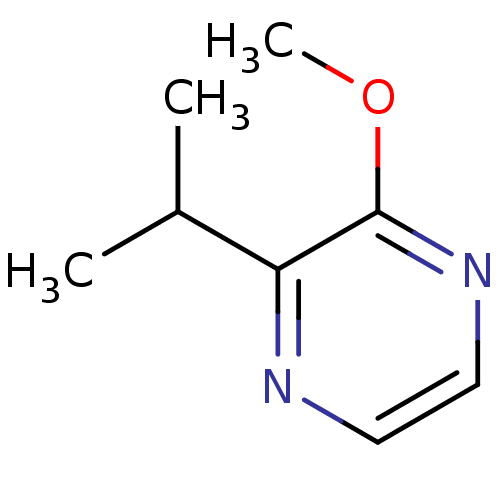
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||