

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA) | ||
| Syringe Reactant: | BDBM18372 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 09/17/08 | ||
| ΔG°: | -10.8± (kcal/mole) | ||
| pH: | 7.2±n/a | ||
| Log10Kb: | 7.77± n/a | ||
| Temperature: | 303.15±n/a (K) | ||
| ΔH° : | n/a | ||
| ΔHobs : | -13.2± (kcal/mole) | ||
| Ionic Strength: | n/a | ||
| no | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | -0.0100± (kcal/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Cywin, CL; Sarver, RW; Klunder, JM; Bills, E; Hoermann, M; Bolton, G; Bratton, LD; Brickwood, JR; Caspers, NL; David, E; Dunbar, JB; Grob, PM; Schwartz, R; Harris, MS; Pauletti, D; Hutchings, RH; Barringer, KJ; Kennedy, RM; Shih, CK; Larsen, SD; Pavlovsky, A; Sorge, CL; Erickson, DA; Pfefferkorn, JA; Joseph, DP; Bainbridge, G; Hattox, SE Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Med Chem51:3804-13 (2008) [PubMed] Article Cywin, CL; Sarver, RW; Klunder, JM; Bills, E; Hoermann, M; Bolton, G; Bratton, LD; Brickwood, JR; Caspers, NL; David, E; Dunbar, JB; Grob, PM; Schwartz, R; Harris, MS; Pauletti, D; Hutchings, RH; Barringer, KJ; Kennedy, RM; Shih, CK; Larsen, SD; Pavlovsky, A; Sorge, CL; Erickson, DA; Pfefferkorn, JA; Joseph, DP; Bainbridge, G; Hattox, SE Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Med Chem51:3804-13 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA) | |||
| Source: | HMGR was expressed as an N-terminal His6 protein truncated from amino acid 441-875 with the mutation M485I in the Escherichia coli. | ||
| Purity: | n/a | ||
| Prep. Method: | Protein was purified via Ni-affinity and gel filtration chromatography. | ||
| Name: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | ||
| Synonyms: | 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA) | HMDH_HUMAN | HMG-CoA Reductase | HMG-CoA reductase (HMGR) | HMGCR | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 97477.10 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P04035 | ||
| Residue: | 888 | ||
| Sequence: |
| ||
| BDBM18372 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | To minimize heat of dilution effects resulting from differences in buffer composition between ligand and protein, ligands were dissolved in dialysate buffer from the final step in the HMGR purification. | ||
| Name | BDBM18372 | ||
| Synonyms: | (3R,5S,6E)-7-[4-(4-fluorophenyl)-2-(N-methylmethanesulfonamido)-6-(propan-2-yl)pyrimidin-5-yl]-3,5-dihydroxyhept-6-enoic acid | CHEMBL1496 | Ros | Rosuvastatin | US9102656, Rosuvastatin | ZD4522 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H28FN3O6S | ||
| Mol. Mass. | 481.538 | ||
| SMILES | CC(C)c1nc(nc(-c2ccc(F)cc2)c1\C=C\[C@@H](O)C[C@@H](O)CC(O)=O)N(C)S(C)(=O)=O |r| | ||
| Structure | 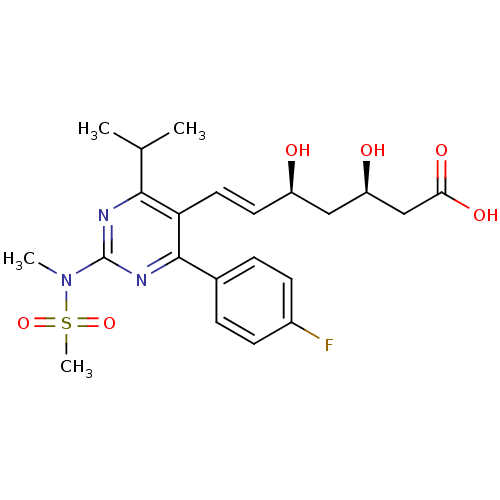
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||