

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | DNA Gyrase | ||
| Syringe Reactant: | BDBM285 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 03/31/03 | ||
| ΔG°: | -35.1±0.200 (kJ/mole) | ||
| pH: | 7.5±n/a | ||
| Log10Kb: | 6.15± 5 | ||
| Temperature: | 298.15±n/a (K) | ||
| ΔH° : | -18±0.5 (kJ/mole) | ||
| ΔHobs : | -18±0.5 (kJ/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | -0.62±0.06 (kJ/mole) | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.0574±0.00100 (kJ/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Lafitte, D; Lamour, V; Tsvetkov, PO; Makarov, AA; Klich, M; Deprez, P; Moras, D; Briand, C; Gilli, R DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5'-methyl group of the noviose. Biochemistry41:7217-23 (2002) [PubMed] Article Lafitte, D; Lamour, V; Tsvetkov, PO; Makarov, AA; Klich, M; Deprez, P; Moras, D; Briand, C; Gilli, R DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5'-methyl group of the noviose. Biochemistry41:7217-23 (2002) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| DNA Gyrase | |||
| Source: | Bacterial clone | ||
| Purity: | 97% | ||
| Prep. Method: | Gilbert, E. J., and Maxwell, A. (1994) Mol. Microbiol. 12, 365-373. | ||
| Name: | DNA gyrase subunit B | ||
| Synonyms: | gyrB | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 89941.28 | ||
| Organism: | Escherichia coli | ||
| Description: | C3SLN3 | ||
| Residue: | 804 | ||
| Sequence: |
| ||
| BDBM285 | |||
| Source: | Synthesis | ||
| Purity: | n/a | ||
| Prep. Method: | Ferroud, D., Collard, J., Klich, M., Dupuis-Hamelin, C., Mauvais, P., Lassaigne, P., Bonnefoy, A., and Musicki, B.(1999) Bioorg. Med. Chem. Lett. 9, 2881-2886. | ||
| Name | BDBM285 | ||
| Synonyms: | Coumarin-Based, DNA Gyrase Inhibitor | RU 64136 | {[7-({3-hydroxy-5-methoxy-6-methyl-4-[(5-methyl-1H-pyrrol-2-yl)carbonyloxy]oxan-2-yl}oxy)-8-methyl-2-oxo-2H-chromen-4-yl]methyl}phosphonate | ||
| Type | Antibiotic | ||
| Emp. Form. | C24H26NO11P | ||
| Mol. Mass. | 535.4382 | ||
| SMILES | COC1C(C)OC(Oc2ccc3c(CP([O-])([O-])=O)cc(=O)oc3c2C)C(O)C1OC(=O)c1ccc(C)[nH]1 | ||
| Structure | 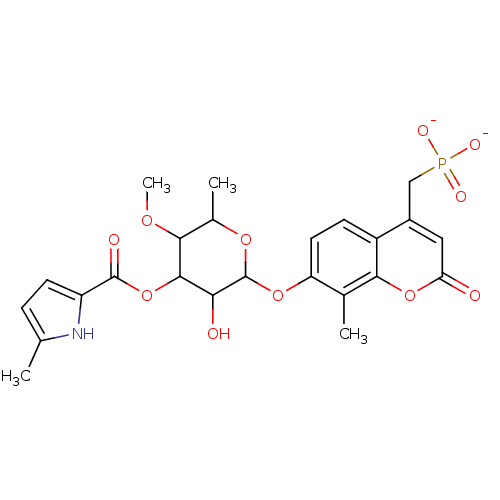
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||