

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | BDBM520 | ||
| Syringe Reactant: | HIV-1 Protease | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 04/20/04 | ||
| ΔG°: | -13.5± (kcal/mole) | ||
| pH: | 5±n/a | ||
| Log10Kb: | n/a | ||
| Temperature: | 298.15±n/a (K) | ||
| ΔH° : | -2.30±0.300 (kcal/mole) | ||
| ΔHobs : | -2.32± (kcal/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | 0.2 | ||
| ΔCp : | -0.38±0.03 (kcal/mole) | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.0400± (kcal/mole-K) | ||
| Comments: | Because of its low solubility, the binding of ritonavir was measured by titrating from 200 uM protease solution into 20 uM ritonavir in the calorimetric cell. | ||
| Citation |  Todd, MJ; Luque, I; Velázquez-Campoy, A; Freire, E Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry39:11876-83 (2000) [PubMed] Article Todd, MJ; Luque, I; Velázquez-Campoy, A; Freire, E Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry39:11876-83 (2000) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| BDBM520 | |||
| Source: | Purified from commercial capsules | ||
| Purity: | n/a | ||
| Prep. Method: | Further purified by HPLC using a semipreparative C-18 reversed-phase column developed with 0-100% acetonitrile in 0.05% TFA. | ||
| Name | BDBM520 | ||
| Synonyms: | 1,3-thiazol-5-ylmethyl N-[(2S,3S,5S)-3-hydroxy-5-[(2S)-3-methyl-2-{[methyl({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl})carbamoyl]amino}butanamido]-1,6-diphenylhexan-2-yl]carbamate | ABT-538 | CHEMBL163 | Norvir | RTV | Ritonavir | ||
| Type | Small organic molecule | ||
| Emp. Form. | C37H48N6O5S2 | ||
| Mol. Mass. | 720.944 | ||
| SMILES | CC(C)[C@H](NC(=O)N(C)Cc1csc(n1)C(C)C)C(=O)N[C@H](C[C@H](O)[C@H](Cc1ccccc1)NC(=O)OCc1cncs1)Cc1ccccc1 | ||
| Structure | 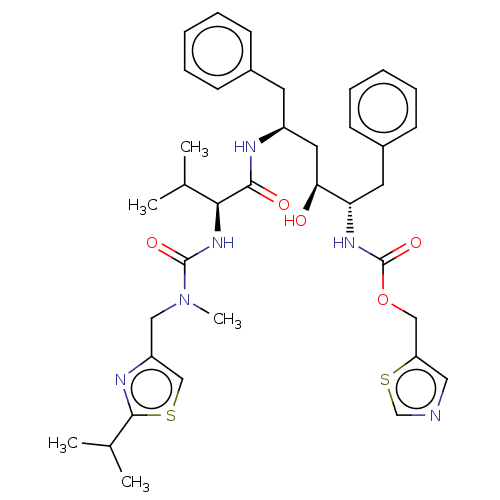
| ||
| HIV-1 Protease | |||
| Source: | Plasmid-encoded protease (Q7K/L33I/L63I ) expressed in Escherichia coli 1458 | ||
| Purity: | 99% | ||
| Prep. Method: | HIV-1 protease was purified and refolded from E. coli inclusion bodies. | ||
| Name: | HIV-1 Protease | ||
| Synonyms: | HIV-1 Protease Triple Mutant | HIV-1 protease pseudo wildtype | ||
| Type: | Protein Complex | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | HIV-1 Protease Mutant (Q7K/L33I/L63I) chain A | ||
| Synonyms: | n/a | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 10782.21 | ||
| Organism: | Human immunodeficiency virus type 1 | ||
| Description: | Using plasmid-encoded mutant protease (Q7K/L33I/L63I designed to remove three hypersensitive autolytic sites) for the stability of protease. | ||
| Residue: | 99 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | HIV-1 Protease Mutant (Q7K/L33I/L63I) chain B | ||
| Synonyms: | n/a | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 10796.24 | ||
| Organism: | Human immunodeficiency virus type 1 | ||
| Description: | Using plasmid-encoded mutant protease (Q7K/L33I/L63I designed to remove three hypersensitive autolytic sites) for the stability of protease. | ||
| Residue: | 99 | ||
| Sequence: |
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||