

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Trypsin | ||
| Syringe Reactant: | BDBM775 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 06/15/04 | ||
| ΔG°: | -6.12±0.0200 (kcal/mole) | ||
| pH: | 8±n/a | ||
| Log10Kb: | 4.40± 3.00 | ||
| Temperature: | 303.35±n/a (K) | ||
| ΔH° : | -3.76±0.140 (kcal/mole) | ||
| ΔHobs : | -3.76±0.140 (kcal/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.0100±0 (kcal/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Talhout, R; Villa, A; Mark, AE; Engberts, JB Understanding binding affinity: a combined isothermal titration calorimetry/molecular dynamics study of the binding of a series of hydrophobically modified benzamidinium chloride inhibitors to trypsin. J Am Chem Soc125:10570-9 (2003) [PubMed] Article Talhout, R; Villa, A; Mark, AE; Engberts, JB Understanding binding affinity: a combined isothermal titration calorimetry/molecular dynamics study of the binding of a series of hydrophobically modified benzamidinium chloride inhibitors to trypsin. J Am Chem Soc125:10570-9 (2003) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| Trypsin | |||
| Source: | no | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Serine protease 1 | ||
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 25790.52 | ||
| Organism: | Bos taurus (bovine) | ||
| Description: | P00760 | ||
| Residue: | 246 | ||
| Sequence: |
| ||
| BDBM775 | |||
| Source: | Synthesized | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM775 | ||
| Synonyms: | [amino(4-propylphenyl)methylidene]azanium | benzamidine deriv. | n-propylbenzamidinium chloride | ||
| Type | n/a | ||
| Emp. Form. | C10H15N2 | ||
| Mol. Mass. | 163.239 | ||
| SMILES | CCCc1ccc(cc1)C(N)=[NH2+] | ||
| Structure | 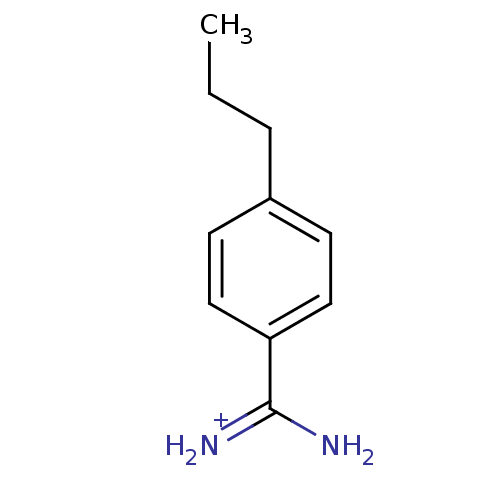
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||