

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Trypsin | ||
| Syringe Reactant: | BDBM776 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 06/15/04 | ||
| ΔG°: | -5.43±0.0500 (kcal/mole) | ||
| pH: | 8±n/a | ||
| Log10Kb: | 3.82± 2.70 | ||
| Temperature: | 310.15±n/a (K) | ||
| ΔH° : | -2.87±0.120 (kcal/mole) | ||
| ΔHobs : | -2.87±0.120 (kcal/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.0100±0 (kcal/mole-K) | ||
| Comments: | Since the inhibitor contains some NH4Cl, control experiment was performed with 10 wt % NH4Cl added to a benzamidinium chloride solution at the usual concentration. Within error, the thermodynamic parameters were identical. Therefore, the concentration of inhibitor, based on weight, can be safely corrected for the amount of NH4Cl present. | ||
| Citation |  Talhout, R; Villa, A; Mark, AE; Engberts, JB Understanding binding affinity: a combined isothermal titration calorimetry/molecular dynamics study of the binding of a series of hydrophobically modified benzamidinium chloride inhibitors to trypsin. J Am Chem Soc125:10570-9 (2003) [PubMed] Article Talhout, R; Villa, A; Mark, AE; Engberts, JB Understanding binding affinity: a combined isothermal titration calorimetry/molecular dynamics study of the binding of a series of hydrophobically modified benzamidinium chloride inhibitors to trypsin. J Am Chem Soc125:10570-9 (2003) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| Trypsin | |||
| Source: | no | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Serine protease 1 | ||
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 25790.52 | ||
| Organism: | Bos taurus (bovine) | ||
| Description: | P00760 | ||
| Residue: | 246 | ||
| Sequence: |
| ||
| BDBM776 | |||
| Source: | Synthesized | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM776 | ||
| Synonyms: | benzamidine deriv. | isopropylbenzamidinium chloride | {amino[4-(propan-2-yl)phenyl]methylidene}azanium | ||
| Type | n/a | ||
| Emp. Form. | C10H15N2 | ||
| Mol. Mass. | 163.239 | ||
| SMILES | CC(C)c1ccc(cc1)C(N)=[NH2+] | ||
| Structure | 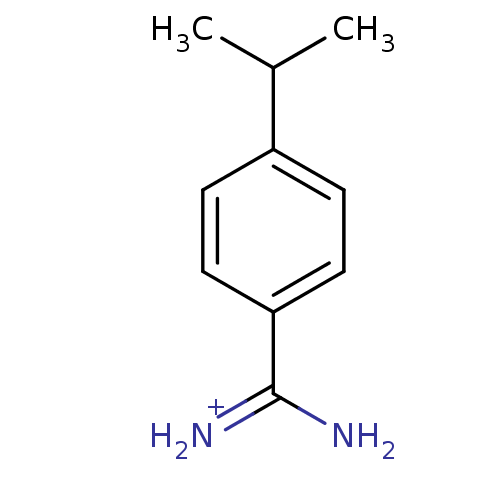
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||