

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Bromodomain-containing protein 4 (BRD4) | ||
| Syringe Reactant: | BDBM50365262 | ||
| Meas. Tech.: | Enzyme Inhibition | ||
| Entry Date: | 12/15/16 | ||
| ΔG°: | -10.1± (kcal/mole) | ||
| pH: | n/a | ||
| Log10Kb: | 7.42± n/a | ||
| Temperature: | 298.15±0 (K) | ||
| ΔH° : | -6.81±0.0700 (kcal/mole) | ||
| ΔHobs: | n/a | ||
| Ionic Strength: | n/a | ||
| n/a | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.0100±0 (kcal/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Tanaka, M; Roberts, JM; Seo, HS; Souza, A; Paulk, J; Scott, TG; DeAngelo, SL; Dhe-Paganon, S; Bradner, JE Design and characterization of bivalent BET inhibitors. Nat Chem Biol12:1089-1096 (2016) [PubMed] Article Tanaka, M; Roberts, JM; Seo, HS; Souza, A; Paulk, J; Scott, TG; DeAngelo, SL; Dhe-Paganon, S; Bradner, JE Design and characterization of bivalent BET inhibitors. Nat Chem Biol12:1089-1096 (2016) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data | ||
| Bromodomain-containing protein 4 (BRD4) | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Bromodomain-containing protein 4 [44-168] | ||
| Synonyms: | BET bromodomain 4(1) (BRD4(1)) | BRD4 | BRD4-BD1 protein (aa 44-168) | BRD4_HUMAN | Bromodomain protein 4 (BRD4-BD1) | Bromodomain-containing protein 4 (BRD4-BD1) | Bromodomain-containing protein 4 (BRD4) (aa 44-168) | Bromodomain-containing protein 4 (BRD4-BD1) | Bromodomain-containing protein 4 bromodomain 1 (BRD4-BD1) | HUNK1 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 14866.49 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | aa (44-168) | ||
| Residue: | 125 | ||
| Sequence: |
| ||
| BDBM50365262 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM50365262 | ||
| Synonyms: | (+)-JQ1 | (S)-JQ1 (1) | CHEMBL1957266 | JQ1 | US10124009, Compound (S)-JQ1 | US10202360, Example JQ-1 | US10308662, Compound JQ-1 | US10407441, Compound (S)-JQ1 | US10617680, Example JQ-1 | US10881668, Compound JQ1 | US10925881, Name (S)-JQ1 | US11020380, Example JQ-1 | US11078188, Example (+)-JQ1 | US11279703, TABLE 6.180 | US11427593, Compound JQ1 | US11466034, Example (+)-JQ1 | US9320741, (S)-JQ1 | US9695172, JQ1 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H25ClN4O2S | ||
| Mol. Mass. | 456.988 | ||
| SMILES | Cc1nnc2[C@H](CC(=O)OC(C)(C)C)N=C(c3c(C)c(C)sc3-n12)c1ccc(Cl)cc1 |r,c:14| | ||
| Structure | 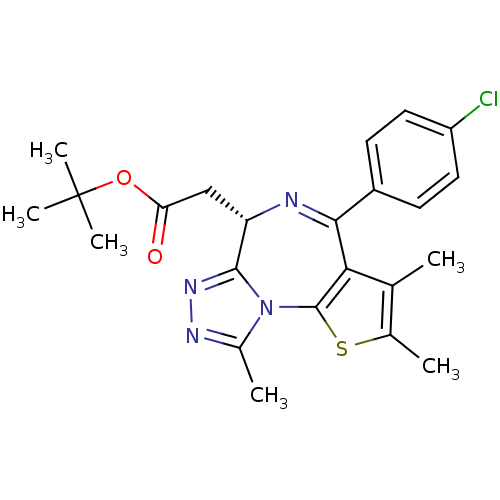
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||