

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Thiol:disulfide interchange protein (DsbA) | ||
| Syringe Reactant: | BDBM231642 | ||
| Meas. Tech.: | Enzyme Inhibition | ||
| Entry Date: | 06/06/17 | ||
| ΔG°: | -57.1± (kJ/mole) | ||
| pH: | 7.4±0 | ||
| Log10Kb: | n/a | ||
| Temperature: | 298.15±0 (K) | ||
| ΔH° : | -43.1±4 (kJ/mole) | ||
| ΔHobs: | n/a | ||
| Ionic Strength: | n/a | ||
| n/a | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.047±0 (kJ/mole-K) | ||
| Comments: | Oxidized AbDsbA | ||
| Citation |  Premkumar, L; Kurth, F; Duprez, W; Gr°ftehauge, MK; King, GJ; Halili, MA; Heras, B; Martin, JL Structure of the Acinetobacter baumannii dithiol oxidase DsbA bound to elongation factor EF-Tu reveals a novel protein interaction site. J Biol Chem289:19869-80 (2014) [PubMed] Article Premkumar, L; Kurth, F; Duprez, W; Gr°ftehauge, MK; King, GJ; Halili, MA; Heras, B; Martin, JL Structure of the Acinetobacter baumannii dithiol oxidase DsbA bound to elongation factor EF-Tu reveals a novel protein interaction site. J Biol Chem289:19869-80 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data | ||
| Thiol:disulfide interchange protein (DsbA) | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Thiol:disulfide interchange protein [27-205] | ||
| Synonyms: | F0KIP0_ACICP | Thiol:disulfide interchange protein (DsbA) | dsbA | ||
| Type: | Protein | ||
| Mol. Mass.: | 20389.27 | ||
| Organism: | Acinetobacter baumannii (AYE strain) | ||
| Description: | A. baumannii DsbA lacking the predicted periplasmic leader sequence (27-205 aa) | ||
| Residue: | 179 | ||
| Sequence: |
| ||
| BDBM231642 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM231642 | ||
| Synonyms: | PSCGPGL | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H43N7O9S | ||
| Mol. Mass. | 629.726 | ||
| SMILES | CC(C)C[C@H](NC(=O)CNC(=O)[C@@H]1CCCN1C(=O)CNC(=O)[C@H](CS)NC(=O)[C@H](CO)NC(=O)[C@@H]1CCCN1)C(O)=O |r| | ||
| Structure | 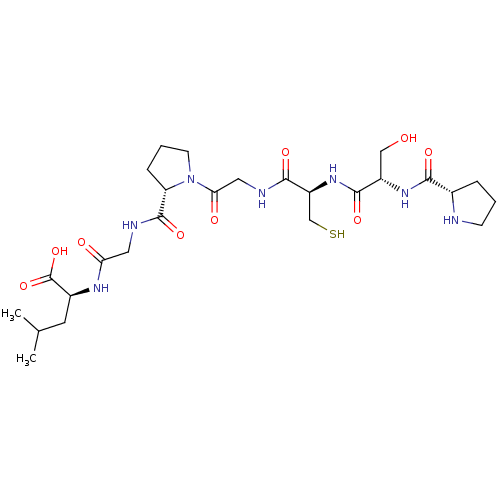
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||