

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Reverse transcriptase (HIV-1 RT)(1-350) | ||
| Syringe Reactant: | BDBM2337 | ||
| Meas. Tech.: | Enzyme Inhibition | ||
| Entry Date: | 06/16/17 | ||
| ΔG°: | -34.7±10.7 (kcal/mole) | ||
| pH: | n/a | ||
| Log10Kb: | n/a | ||
| Temperature: | 298.15±0 (K) | ||
| ΔH° : | -55.0± (kcal/mole) | ||
| ΔHobs: | n/a | ||
| Ionic Strength: | n/a | ||
| n/a | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | -0.0200±0.0100 (kcal/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Decha, P; Intharathep, P; Udommaneethanakit, T; Sompornpisut, P; Hannongbua, S; Wolschann, P; Parasuk, V Theoretical studies on the molecular basis of HIV-1RT/NNRTIs interactions. J Enzyme Inhib Med Chem26:29-36 (2011) [PubMed] Article Decha, P; Intharathep, P; Udommaneethanakit, T; Sompornpisut, P; Hannongbua, S; Wolschann, P; Parasuk, V Theoretical studies on the molecular basis of HIV-1RT/NNRTIs interactions. J Enzyme Inhib Med Chem26:29-36 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data | ||
| Reverse transcriptase (HIV-1 RT)(1-350) | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Reverse transcriptase (HIV-1 RT)(aa 1-350) | ||
| Synonyms: | POL_HV1H2 | gag-pol | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 39217.95 | ||
| Organism: | Human immunodeficiency virus type 1 | ||
| Description: | HIV-1 RT variant (1-350 aa) | ||
| Residue: | 350 | ||
| Sequence: |
| ||
| BDBM2337 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM2337 | ||
| Synonyms: | 6-benzyl-1-(ethoxymethyl)-5-(propan-2-yl)-1,2,3,4-tetrahydropyrimidine-2,4-dione | CHEMBL35033 | Coactinon | Emivirine | Emivirine (EMV) | HEPT deriv. | I-EBU | MKC-442 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H22N2O3 | ||
| Mol. Mass. | 302.3682 | ||
| SMILES | CCOCn1c(Cc2ccccc2)c(C(C)C)c(=O)[nH]c1=O | ||
| Structure | 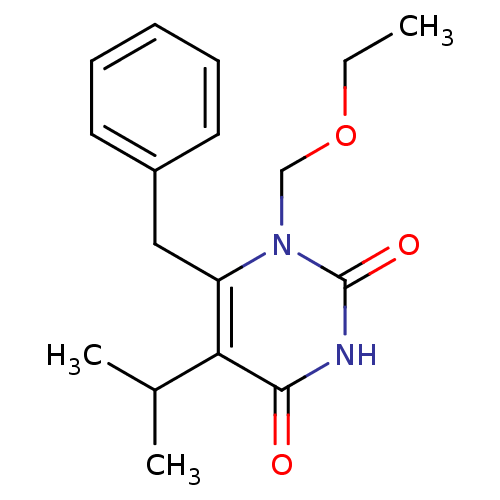
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||