

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Oxysterols receptor LXR-beta | ||
| Ligand | BDBM304697 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | LXR alpha/beta Radioligand Binding Assay | ||
| Ki | 6.00±n/a nM | ||
| Citation |  Claremon, DA; Dong, C; Fan, Y; Leftheris, K; Lotesta, SD; Singh, SB; Tice, CM; Zhao, W; Zheng, Y; Zhuang, L Piperazine derivatives as liver X receptor modulators US Patent US10144715 Publication Date 12/4/2018 Claremon, DA; Dong, C; Fan, Y; Leftheris, K; Lotesta, SD; Singh, SB; Tice, CM; Zhao, W; Zheng, Y; Zhuang, L Piperazine derivatives as liver X receptor modulators US Patent US10144715 Publication Date 12/4/2018 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Oxysterols receptor LXR-beta | |||
| Name: | Oxysterols receptor LXR-beta | ||
| Synonyms: | LXRB | Liver X receptor beta (NR1H2) | Liver X, LXR beta | NER | NR1H2 | NR1H2_HUMAN | Nuclear receptor NER | UNR | Ubiquitously-expressed nuclear receptor | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 50978.79 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P55055 | ||
| Residue: | 460 | ||
| Sequence: |
| ||
| BDBM304697 | |||
| n/a | |||
| Name | BDBM304697 | ||
| Synonyms: | 4-((S)-3-isopropyl-4-(((1s,4R)-4-(trifluoromethyl)cyclohexyl)methyl)piperazin-1-yl)-2-(methylsulfonyl)benzonitrile | US10144715, Compound 50-1 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H32F3N3O2S | ||
| Mol. Mass. | 471.579 | ||
| SMILES | CC(C)[C@H]1CN(CCN1C[C@H]1CC[C@H](CC1)C(F)(F)F)c1ccc(C#N)c(c1)S(C)(=O)=O |r,wU:10.10,13.17,wD:3.2,(5.51,-26.58,;4.17,-25.81,;2.84,-26.58,;4.17,-24.27,;5.51,-23.5,;5.51,-21.96,;4.17,-21.19,;2.84,-21.96,;2.84,-23.5,;1.51,-24.27,;.17,-23.5,;-1.16,-24.27,;-2.5,-23.5,;-2.5,-21.96,;-1.16,-21.19,;.17,-21.96,;-3.83,-21.19,;-5.16,-21.96,;-3.83,-19.65,;-3.83,-22.73,;6.84,-21.19,;8.17,-21.96,;9.51,-21.19,;9.54,-19.62,;10.88,-18.87,;12.23,-18.11,;8.18,-18.84,;6.84,-19.65,;8.17,-17.3,;9.71,-17.28,;8.15,-15.76,;6.63,-17.32,)| | ||
| Structure | 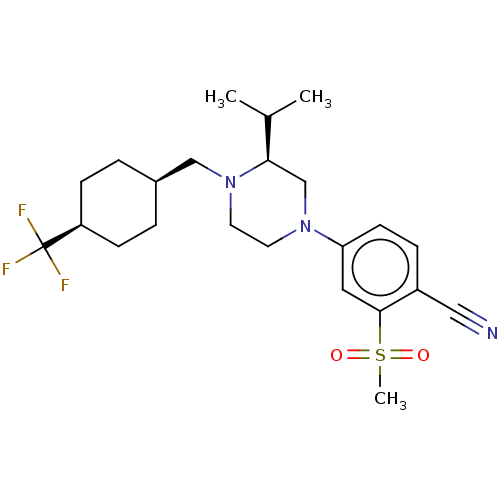
| ||