| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Carbonic anhydrase 2 |
|---|
| Ligand | BDBM11638 |
|---|
| Substrate/Competitor | BDBM10856 |
|---|
| Meas. Tech. | CA Inhibition Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Ki | 40±n/a nM |
|---|
| Citation |  Menchise, V; De Simone, G; Alterio, V; Di Fiore, A; Pedone, C; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors: stacking with Phe131 determines active site binding region of inhibitors as exemplified by the X-ray crystal structure of a membrane-impermeant antitumor sulfonamide complexed with isozyme II. J Med Chem48:5721-7 (2005) [PubMed] Article Menchise, V; De Simone, G; Alterio, V; Di Fiore, A; Pedone, C; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors: stacking with Phe131 determines active site binding region of inhibitors as exemplified by the X-ray crystal structure of a membrane-impermeant antitumor sulfonamide complexed with isozyme II. J Med Chem48:5721-7 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Carbonic anhydrase 2 |
|---|
| Name: | Carbonic anhydrase 2 |
|---|
| Synonyms: | CA-II | CA2 | CAC | CAH2_HUMAN | Carbonate dehydratase II | Carbonic anhydrase 2 (CA II) | Carbonic anhydrase 2 (CA-II) | Carbonic anhydrase 2 (Recombinant CA II) | Carbonic anhydrase C | Carbonic anhydrase II (CA II) | Carbonic anhydrase II (CA-II) | Carbonic anhydrase II (CAII) | Carbonic anhydrase II (hCA II) | Carbonic anhydrase isoenzyme II (hCA II) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 29250.71 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P00918 |
|---|
| Residue: | 260 |
|---|
| Sequence: | MSHHWGYGKHNGPEHWHKDFPIAKGERQSPVDIDTHTAKYDPSLKPLSVSYDQATSLRIL
NNGHAFNVEFDDSQDKAVLKGGPLDGTYRLIQFHFHWGSLDGQGSEHTVDKKKYAAELHL
VHWNTKYGDFGKAVQQPDGLAVLGIFLKVGSAKPGLQKVVDVLDSIKTKGKSADFTNFDP
RGLLPESLDYWTYPGSLTTPPLLECVTWIVLKEPISVSSEQVLKFRKLNFNGEGEPEELM
VDNWRPAQPLKNRQIKASFK
|
|
|
|---|
| BDBM11638 |
|---|
| BDBM10856 |
|---|
| Name | BDBM11638 |
|---|
| Synonyms: | CHEMBL26 | Compound 7 | N-[(1-ethylpyrrolidin-2-yl)methyl]-2-methoxy-5-sulfamoylbenzamide | SULPIRIDE,(+) | SULPIRIDE,(-) | Sulpiride | Sulpiride, SLP | Sulpiride-R | US10172837, Sulpiride |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H23N3O4S |
|---|
| Mol. Mass. | 341.426 |
|---|
| SMILES | CCN1CCCC1CNC(=O)c1cc(ccc1OC)S(N)(=O)=O |
|---|
| Structure | 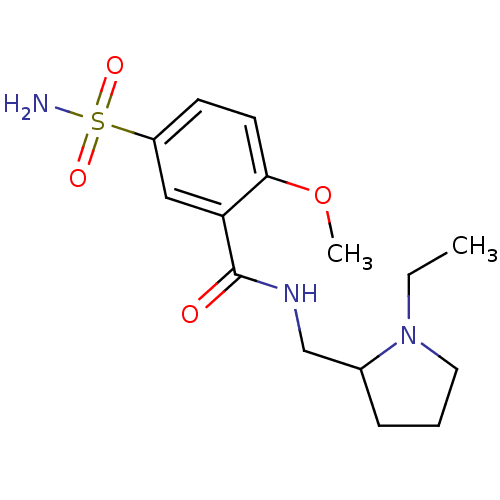
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Menchise, V; De Simone, G; Alterio, V; Di Fiore, A; Pedone, C; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors: stacking with Phe131 determines active site binding region of inhibitors as exemplified by the X-ray crystal structure of a membrane-impermeant antitumor sulfonamide complexed with isozyme II. J Med Chem48:5721-7 (2005) [PubMed] Article
Menchise, V; De Simone, G; Alterio, V; Di Fiore, A; Pedone, C; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors: stacking with Phe131 determines active site binding region of inhibitors as exemplified by the X-ray crystal structure of a membrane-impermeant antitumor sulfonamide complexed with isozyme II. J Med Chem48:5721-7 (2005) [PubMed] Article