| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase [1-303] |
|---|
| Ligand | BDBM65495 |
|---|
| Substrate/Competitor | BDBM65498 |
|---|
| Meas. Tech. | NAD glycohydrolase assay |
|---|
| pH | 7.4±0 |
|---|
| Temperature | 310.15±0 K |
|---|
| IC50 | 88000±8000 nM |
|---|
| Citation |  Muller-Steffner, H; Jacques, SA; Kuhn, I; Schultz, MD; Botta, D; Osswald, P; Maechling, C; Lund, FE; Kellenberger, E Efficient Inhibition of SmNACE by Coordination Complexes Is Abolished by S. mansoni Sequestration of Metal. ACS Chem Biol12:1787-1795 (2017) [PubMed] Article Muller-Steffner, H; Jacques, SA; Kuhn, I; Schultz, MD; Botta, D; Osswald, P; Maechling, C; Lund, FE; Kellenberger, E Efficient Inhibition of SmNACE by Coordination Complexes Is Abolished by S. mansoni Sequestration of Metal. ACS Chem Biol12:1787-1795 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase [1-303] |
|---|
| Name: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase [1-303] |
|---|
| Synonyms: | C-His6-SmNACE |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 35588.81 |
|---|
| Organism: | Schistosoma mansoni |
|---|
| Description: | Q32TF5 6 His tagged |
|---|
| Residue: | 309 |
|---|
| Sequence: | MMNVILFLTLSNIFVFNSAQHQINLLSEIVQSRCTQWKVEHGATNISCSEIWNSFESILL
STHTKSACVMKSGLFDDFVYQLFELEQQQQQRHHTIQTEQYFHSQVMNIIRGMCKRLGVC
RSLETTFPGYLFDELNWCNGSLTGNTKYGTVCGCDYKSNVVHAFWQSASAEYARRASGNI
FVVLNGSVKAPFNENKTFGKIELPLLKHPRVQQLTVKLVHSLEDVNNRQTCESWSLQELA
NKLNSVHIPFRCIDDPLEFRHYQCIENPGKQLCQFSASTRSNVETLLILFPLVICLTFYT
SMNHHHHHH
|
|
|
|---|
| BDBM65495 |
|---|
| BDBM65498 |
|---|
| Name | BDBM65495 |
|---|
| Synonyms: | SmNACE inhibitor, 1 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H12N2O4 |
|---|
| Mol. Mass. | 260.2454 |
|---|
| SMILES | Cc1occc1C(=O)N\N=C\c1ccc(O)c(O)c1 |
|---|
| Structure | 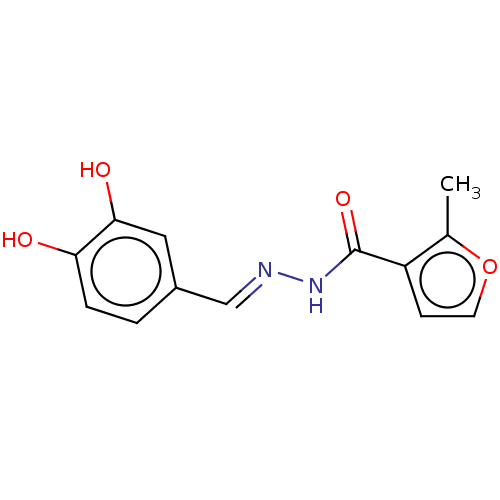
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Muller-Steffner, H; Jacques, SA; Kuhn, I; Schultz, MD; Botta, D; Osswald, P; Maechling, C; Lund, FE; Kellenberger, E Efficient Inhibition of SmNACE by Coordination Complexes Is Abolished by S. mansoni Sequestration of Metal. ACS Chem Biol12:1787-1795 (2017) [PubMed] Article
Muller-Steffner, H; Jacques, SA; Kuhn, I; Schultz, MD; Botta, D; Osswald, P; Maechling, C; Lund, FE; Kellenberger, E Efficient Inhibition of SmNACE by Coordination Complexes Is Abolished by S. mansoni Sequestration of Metal. ACS Chem Biol12:1787-1795 (2017) [PubMed] Article