| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dipeptidyl peptidase 9 |
|---|
| Ligand | BDBM12642 |
|---|
| Substrate/Competitor | BDBM11057 |
|---|
| Meas. Tech. | DPPIV Inhibition Assay |
|---|
| Ki | >30000±n/a nM |
|---|
| Citation |  Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Dipeptidyl peptidase 9 |
|---|
| Name: | Dipeptidyl peptidase 9 |
|---|
| Synonyms: | DPP9 | DPP9_HUMAN | DPRP-2 | DPRP2 | Dipeptidyl peptidase 9 (DDP9) | Dipeptidyl peptidase 9 (DPP-9) | Dipeptidyl peptidase 9 (DPP9) | Dipeptidyl peptidase IV-related protein 2 | Dipeptidyl peptidase IX | Dipeptidyl peptidase IX (DDP-IX) | Dipeptidyl peptidase-like protein 9 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 98260.70 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q86TI2 |
|---|
| Residue: | 863 |
|---|
| Sequence: | MATTGTPTADRGDAAATDDPAARFQVQKHSWDGLRSIIHGSRKYSGLIVNKAPHDFQFVQ
KTDESGPHSHRLYYLGMPYGSRENSLLYSEIPKKVRKEALLLLSWKQMLDHFQATPHHGV
YSREEELLRERKRLGVFGITSYDFHSESGLFLFQASNSLFHCRDGGKNGFMVSPMKPLEI
KTQCSGPRMDPKICPADPAFFSFINNSDLWVANIETGEERRLTFCHQGLSNVLDDPKSAG
VATFVIQEEFDRFTGYWWCPTASWEGSEGLKTLRILYEEVDESEVEVIHVPSPALEERKT
DSYRYPRTGSKNPKIALKLAEFQTDSQGKIVSTQEKELVQPFSSLFPKVEYIARAGWTRD
GKYAWAMFLDRPQQWLQLVLLPPALFIPSTENEEQRLASARAVPRNVQPYVVYEEVTNVW
INVHDIFYPFPQSEGEDELCFLRANECKTGFCHLYKVTAVLKSQGYDWSEPFSPGEDEFK
CPIKEEIALTSGEWEVLARHGSKIWVNEETKLVYFQGTKDTPLEHHLYVVSYEAAGEIVR
LTTPGFSHSCSMSQNFDMFVSHYSSVSTPPCVHVYKLSGPDDDPLHKQPRFWASMMEAAS
CPPDYVPPEIFHFHTRSDVRLYGMIYKPHALQPGKKHPTVLFVYGGPQVQLVNNSFKGIK
YLRLNTLASLGYAVVVIDGRGSCQRGLRFEGALKNQMGQVEIEDQVEGLQFVAEKYGFID
LSRVAIHGWSYGGFLSLMGLIHKPQVFKVAIAGAPVTVWMAYDTGYTERYMDVPENNQHG
YEAGSVALHVEKLPNEPNRLLILHGFLDENVHFFHTNFLVSQLIRAGKPYQLQIYPNERH
SIRCPESGEHYEVTLLHFLQEYL
|
|
|
|---|
| BDBM12642 |
|---|
| BDBM11057 |
|---|
| Name | BDBM12642 |
|---|
| Synonyms: | (2S,5R)-5-ethynyl-1-[N-(3-hydroxy-1-adamantyl)glycyl]pyrrolidine-2-carbonitrile | (2S,5R)-5-ethynyl-1-{2-[(3-hydroxyadamantan-1-yl)amino]acetyl}pyrrolidine-2-carbonitrile | C5-substituted pyrrolidine 35 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H25N3O2 |
|---|
| Mol. Mass. | 327.4207 |
|---|
| SMILES | OC12CC3CC(C1)CC(C3)(C2)NCC(=O)N1[C@H](CC[C@H]1C#N)C#C |r,TLB:6:1:9:5.4.7,THB:7:5:2:9.10.8,7:8:2:5.4.6,6:5:9:10.2.1,0:1:9:5.4.7| |
|---|
| Structure | 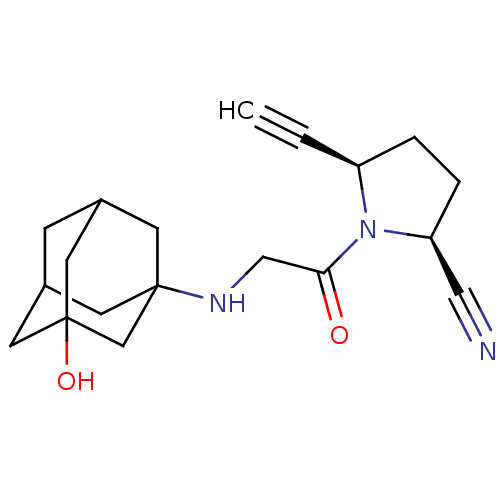
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article
Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article