| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM12661 |
|---|
| Substrate/Competitor | BDBM12679 |
|---|
| Meas. Tech. | Enzyme Assay and Determination of the Inhibition Constants. |
|---|
| Ki | >4200±n/a nM |
|---|
| Citation |  Quan, ML; Lam, PY; Han, Q; Pinto, DJ; He, MY; Li, R; Ellis, CD; Clark, CG; Teleha, CA; Sun, JH; Alexander, RS; Bai, S; Luettgen, JM; Knabb, RM; Wong, PC; Wexler, RR Discovery of 1-(3'-aminobenzisoxazol-5'-yl)-3-trifluoromethyl-N-[2-fluoro-4-[(2'-dimethylaminomethyl)imidazol-1-yl]phenyl]-1H-pyrazole-5-carboxyamide hydrochloride (razaxaban), a highly potent, selective, and orally bioavailable factor Xa inhibitor. J Med Chem48:1729-44 (2005) [PubMed] Article Quan, ML; Lam, PY; Han, Q; Pinto, DJ; He, MY; Li, R; Ellis, CD; Clark, CG; Teleha, CA; Sun, JH; Alexander, RS; Bai, S; Luettgen, JM; Knabb, RM; Wong, PC; Wexler, RR Discovery of 1-(3'-aminobenzisoxazol-5'-yl)-3-trifluoromethyl-N-[2-fluoro-4-[(2'-dimethylaminomethyl)imidazol-1-yl]phenyl]-1H-pyrazole-5-carboxyamide hydrochloride (razaxaban), a highly potent, selective, and orally bioavailable factor Xa inhibitor. J Med Chem48:1729-44 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 25790.52 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | P00760 |
|---|
| Residue: | 246 |
|---|
| Sequence: | MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVS
AAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLN
SRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQIT
SNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIK
QTIASN
|
|
|
|---|
| BDBM12661 |
|---|
| BDBM12679 |
|---|
| Name | BDBM12661 |
|---|
| Synonyms: | 1-(3-Aminobenzisoxazol-5-yl)-3-trifluoromethyl-5-[[4-(2-aminosulfonyl-4-pyridinyl)-3-fluorophenyl]aminocarbonyl]pyrazole, Trifluoroacetic Acid Salt | 1-(3-amino-1,2-benzoxazol-5-yl)-N-[2-fluoro-4-(3-sulfamoylpyridin-4-yl)phenyl]-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide; 2,2,2-trifluoroacetic acid | Razaxaban Analogue | aminobenzisoxazole 23 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H15F4N7O4S |
|---|
| Mol. Mass. | 561.468 |
|---|
| SMILES | Nc1noc2ccc(cc12)-n1nc(cc1C(=O)Nc1ccc(cc1F)-c1ccncc1S(N)(=O)=O)C(F)(F)F |
|---|
| Structure | 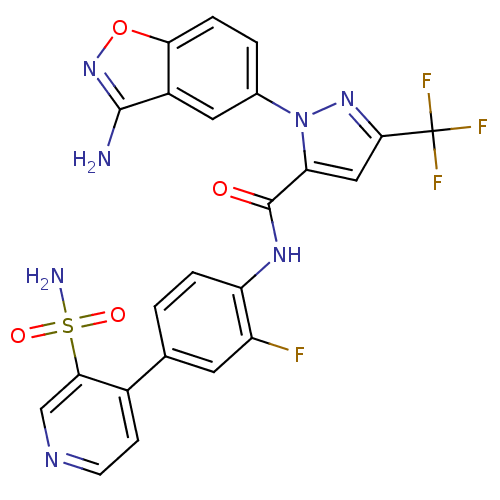
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Quan, ML; Lam, PY; Han, Q; Pinto, DJ; He, MY; Li, R; Ellis, CD; Clark, CG; Teleha, CA; Sun, JH; Alexander, RS; Bai, S; Luettgen, JM; Knabb, RM; Wong, PC; Wexler, RR Discovery of 1-(3'-aminobenzisoxazol-5'-yl)-3-trifluoromethyl-N-[2-fluoro-4-[(2'-dimethylaminomethyl)imidazol-1-yl]phenyl]-1H-pyrazole-5-carboxyamide hydrochloride (razaxaban), a highly potent, selective, and orally bioavailable factor Xa inhibitor. J Med Chem48:1729-44 (2005) [PubMed] Article
Quan, ML; Lam, PY; Han, Q; Pinto, DJ; He, MY; Li, R; Ellis, CD; Clark, CG; Teleha, CA; Sun, JH; Alexander, RS; Bai, S; Luettgen, JM; Knabb, RM; Wong, PC; Wexler, RR Discovery of 1-(3'-aminobenzisoxazol-5'-yl)-3-trifluoromethyl-N-[2-fluoro-4-[(2'-dimethylaminomethyl)imidazol-1-yl]phenyl]-1H-pyrazole-5-carboxyamide hydrochloride (razaxaban), a highly potent, selective, and orally bioavailable factor Xa inhibitor. J Med Chem48:1729-44 (2005) [PubMed] Article