

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Tyrosine-protein kinase ABL1 [201-500,F359V] | ||
| Ligand | BDBM6572 | ||
| Substrate/Competitor | BDBM13531 | ||
| Meas. Tech. | Kinase Assay and Binding Constant Measurement | ||
| Kd | 1±n/a nM | ||
| Citation |  Carter, TA; Wodicka, LM; Shah, NP; Velasco, AM; Fabian, MA; Treiber, DK; Milanov, ZV; Atteridge, CE; Biggs, WH; Edeen, PT; Floyd, M; Ford, JM; Grotzfeld, RM; Herrgard, S; Insko, DE; Mehta, SA; Patel, HK; Pao, W; Sawyers, CL; Varmus, H; Zarrinkar, PP; Lockhart, DJ Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci U S A102:11011-6 (2005) [PubMed] Article Carter, TA; Wodicka, LM; Shah, NP; Velasco, AM; Fabian, MA; Treiber, DK; Milanov, ZV; Atteridge, CE; Biggs, WH; Edeen, PT; Floyd, M; Ford, JM; Grotzfeld, RM; Herrgard, S; Insko, DE; Mehta, SA; Patel, HK; Pao, W; Sawyers, CL; Varmus, H; Zarrinkar, PP; Lockhart, DJ Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci U S A102:11011-6 (2005) [PubMed] Article | ||
| More Info.: | Get all data from this article, Solution Info, Assay Method | ||
| Tyrosine-protein kinase ABL1 [201-500,F359V] | |||
| Name: | Tyrosine-protein kinase ABL1 [201-500,F359V] | ||
| Synonyms: | ABL | ABL Mutant (F359V) | ABL1 | ABL1_HUMAN | JTK7 | Proto-oncogene tyrosine-protein kinase ABL1 mutant F359V | c-ABL | p150 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 34547.72 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P00519[201-500,F359V] | ||
| Residue: | 300 | ||
| Sequence: |
| ||
| BDBM6572 | |||
| BDBM13531 | |||
| Name | BDBM6572 | ||
| Synonyms: | 6-(2,6-dichlorophenyl)-2-[(4-fluoro-3-methylphenyl)amino]-8-methyl-7H,8H-pyrido[2,3-d]pyrimidin-7-one | 6-(2,6-dichlorophenyl)-2-[(4-fluoro-3-methylphenyl)amino]-8-methylpyrido[2,3-d]pyrimidin-7(8H)-one | PD-180970 | PD180970 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H15Cl2FN4O | ||
| Mol. Mass. | 429.274 | ||
| SMILES | Cc1cc(Nc2ncc3cc(-c4c(Cl)cccc4Cl)c(=O)n(C)c3n2)ccc1F |(-2.83,-3.52,;-4.17,-2.76,;-4.17,-1.22,;-5.51,-.46,;-5.51,1.08,;-4.18,1.86,;-4.18,3.4,;-2.84,4.17,;-1.51,3.4,;-.18,4.17,;1.16,3.4,;2.49,4.17,;2.49,5.71,;1.16,6.48,;3.83,6.48,;5.16,5.71,;5.16,4.17,;3.83,3.39,;3.83,1.85,;1.16,1.86,;2.49,1.09,;-.18,1.09,;-.18,-.45,;-1.51,1.86,;-2.84,1.09,;-6.84,-1.23,;-6.83,-2.77,;-5.5,-3.54,;-5.49,-5.08,)| | ||
| Structure | 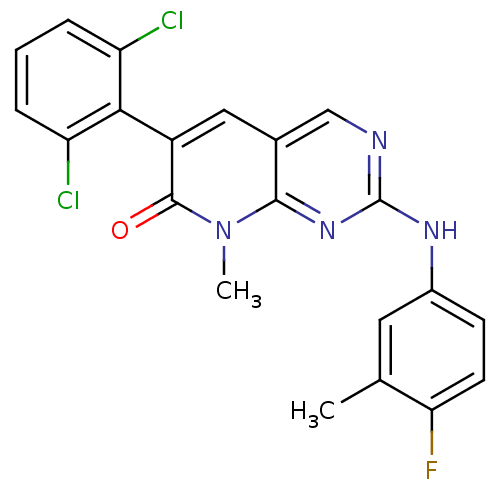
| ||