| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prothrombin |
|---|
| Ligand | BDBM13642 |
|---|
| Substrate/Competitor | BDBM13573 |
|---|
| Meas. Tech. | Thrombin Inhibition Assay |
|---|
| Ki | 4866±n/a nM |
|---|
| Citation |  Matter, H; Will, DW; Nazare, M; Schreuder, H; Laux, V; Wehner, V Structural requirements for factor Xa inhibition by 3-oxybenzamides with neutral P1 substituents: combining X-ray crystallography, 3D-QSAR, and tailored scoring functions. J Med Chem48:3290-312 (2005) [PubMed] Article Matter, H; Will, DW; Nazare, M; Schreuder, H; Laux, V; Wehner, V Structural requirements for factor Xa inhibition by 3-oxybenzamides with neutral P1 substituents: combining X-ray crystallography, 3D-QSAR, and tailored scoring functions. J Med Chem48:3290-312 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Prothrombin |
|---|
| Name: | Prothrombin |
|---|
| Synonyms: | Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 70029.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P00734 |
|---|
| Residue: | 622 |
|---|
| Sequence: | MAHVRGLQLPGCLALAALCSLVHSQHVFLAPQQARSLLQRVRRANTFLEEVRKGNLEREC
VEETCSYEEAFEALESSTATDVFWAKYTACETARTPRDKLAACLEGNCAEGLGTNYRGHV
NITRSGIECQLWRSRYPHKPEINSTTHPGADLQENFCRNPDSSTTGPWCYTTDPTVRRQE
CSIPVCGQDQVTVAMTPRSEGSSVNLSPPLEQCVPDRGQQYQGRLAVTTHGLPCLAWASA
QAKALSKHQDFNSAVQLVENFCRNPDGDEEGVWCYVAGKPGDFGYCDLNYCEEAVEEETG
DGLDEDSDRAIEGRTATSEYQTFFNPRTFGSGEADCGLRPLFEKKSLEDKTERELLESYI
DGRIVEGSDAEIGMSPWQVMLFRKSPQELLCGASLISDRWVLTAAHCLLYPPWDKNFTEN
DLLVRIGKHSRTRYERNIEKISMLEKIYIHPRYNWRENLDRDIALMKLKKPVAFSDYIHP
VCLPDRETAASLLQAGYKGRVTGWGNLKETWTANVGKGQPSVLQVVNLPIVERPVCKDST
RIRITDNMFCAGYKPDEGKRGDACEGDSGGPFVMKSPFNNRWYQMGIVSWGEGCDRDGKY
GFYTHVFRLKKWIQKVIDQFGE
|
|
|
|---|
| BDBM13642 |
|---|
| BDBM13573 |
|---|
| Name | BDBM13642 |
|---|
| Synonyms: | 3-Oxybenzamide 28 | 3-[2-(2,4-dichlorophenyl)ethoxy]-4-methyl-N-{[1-(pyridin-4-yl)piperidin-4-yl]methyl}benzamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H29Cl2N3O2 |
|---|
| Mol. Mass. | 498.444 |
|---|
| SMILES | Cc1ccc(cc1OCCc1ccc(Cl)cc1Cl)C(=O)NCC1CCN(CC1)c1ccncc1 |
|---|
| Structure | 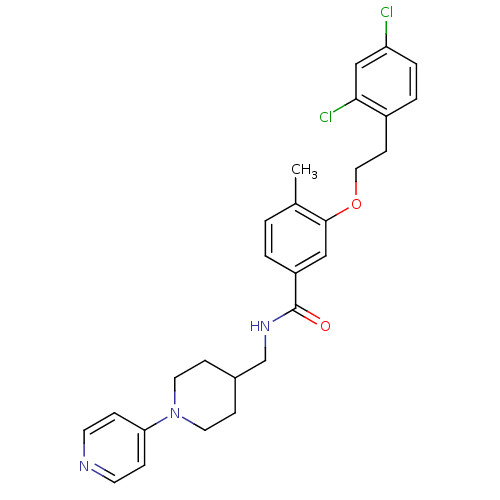
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Matter, H; Will, DW; Nazare, M; Schreuder, H; Laux, V; Wehner, V Structural requirements for factor Xa inhibition by 3-oxybenzamides with neutral P1 substituents: combining X-ray crystallography, 3D-QSAR, and tailored scoring functions. J Med Chem48:3290-312 (2005) [PubMed] Article
Matter, H; Will, DW; Nazare, M; Schreuder, H; Laux, V; Wehner, V Structural requirements for factor Xa inhibition by 3-oxybenzamides with neutral P1 substituents: combining X-ray crystallography, 3D-QSAR, and tailored scoring functions. J Med Chem48:3290-312 (2005) [PubMed] Article