| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma |
|---|
| Ligand | BDBM14392 |
|---|
| Substrate/Competitor | BDBM14391 |
|---|
| Meas. Tech. | PDE Enzyme Inhibitor Assays |
|---|
| pH | 7.4±n/a |
|---|
| Temperature | 303.15±n/a K |
|---|
| IC50 | 386±n/a nM |
|---|
| Comments | cGMP in a 3:1 ratio unlabeled to 3H-labeled at a concentration 1/3Km, such that IC50 = Ki. |
|---|
| Citation |  Allerton, CM; Barber, CG; Beaumont, KC; Brown, DG; Cole, SM; Ellis, D; Lane, CA; Maw, GN; Mount, NM; Rawson, DJ; Robinson, CM; Street, SD; Summerhill, NW A novel series of potent and selective PDE5 inhibitors with potential for high and dose-independent oral bioavailability. J Med Chem49:3581-94 (2006) [PubMed] Article Allerton, CM; Barber, CG; Beaumont, KC; Brown, DG; Cole, SM; Ellis, D; Lane, CA; Maw, GN; Mount, NM; Rawson, DJ; Robinson, CM; Street, SD; Summerhill, NW A novel series of potent and selective PDE5 inhibitors with potential for high and dose-independent oral bioavailability. J Med Chem49:3581-94 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma |
|---|
| Name: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma |
|---|
| Synonyms: | CNRG_CANLF | GMP-PDE gamma | PDE6G | PDEG | Phosphodiesterase Type 6 (PDE6) | Retinal rod rhodopsin-sensitive cGMP 3 ,5 -cyclic phosphodiesterase subunit gamma |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 9656.12 |
|---|
| Organism: | Canis lupus familiaris (Dog) |
|---|
| Description: | n/a |
|---|
| Residue: | 87 |
|---|
| Sequence: | MNLEPPKAEIRSATRVIGGPVTPRKGPPKFKQRQTRQFKSKPPKKGVQGFGDDIPGMEGL
GTDITVICPWEAFNHLELHELAQYGII
|
|
|
|---|
| BDBM14392 |
|---|
| BDBM14391 |
|---|
| Name | BDBM14392 |
|---|
| Synonyms: | 3-ethyl-5-{5-[(4-ethylpiperazine-1-)sulfonyl]-2-(2-methoxyethoxy)pyridin-3-yl}-2-(pyridin-2-ylmethyl)-2H,6H,7H-pyrazolo[4,3-d]pyrimidin-7-one | CHEMBL209157 | Sildenafil Pyridyl Methyl Analogue 1 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H34N8O5S |
|---|
| Mol. Mass. | 582.674 |
|---|
| SMILES | CCN1CCN(CC1)S(=O)(=O)c1cnc(OCCOC)c(c1)-c1nc2c(CC)n(Cc3ccccn3)nc2c(=O)[nH]1 |
|---|
| Structure | 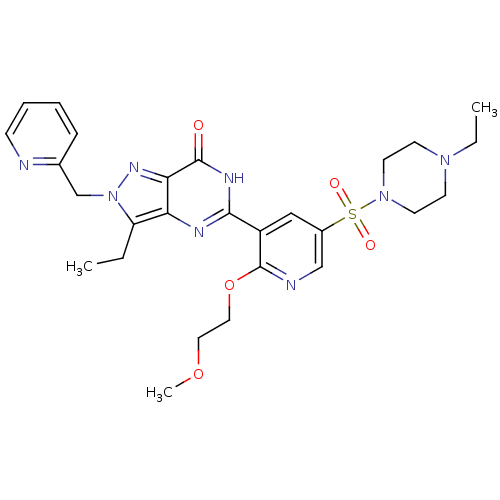
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Allerton, CM; Barber, CG; Beaumont, KC; Brown, DG; Cole, SM; Ellis, D; Lane, CA; Maw, GN; Mount, NM; Rawson, DJ; Robinson, CM; Street, SD; Summerhill, NW A novel series of potent and selective PDE5 inhibitors with potential for high and dose-independent oral bioavailability. J Med Chem49:3581-94 (2006) [PubMed] Article
Allerton, CM; Barber, CG; Beaumont, KC; Brown, DG; Cole, SM; Ellis, D; Lane, CA; Maw, GN; Mount, NM; Rawson, DJ; Robinson, CM; Street, SD; Summerhill, NW A novel series of potent and selective PDE5 inhibitors with potential for high and dose-independent oral bioavailability. J Med Chem49:3581-94 (2006) [PubMed] Article