| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease hepsin |
|---|
| Ligand | BDBM14480 |
|---|
| Substrate/Competitor | BDBM13573 |
|---|
| Meas. Tech. | Enzyme Assay and Determination of the Inhibition Constants |
|---|
| Ki | 2600±n/a nM |
|---|
| Citation |  Katz, BA; Luong, C; Ho, JD; Somoza, JR; Gjerstad, E; Tang, J; Williams, SR; Verner, E; Mackman, RL; Young, WB; Sprengeler, PA; Chan, H; Mortara, K; Janc, JW; McGrath, ME Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA). J Mol Biol344:527-47 (2004) [PubMed] Article Katz, BA; Luong, C; Ho, JD; Somoza, JR; Gjerstad, E; Tang, J; Williams, SR; Verner, E; Mackman, RL; Young, WB; Sprengeler, PA; Chan, H; Mortara, K; Janc, JW; McGrath, ME Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA). J Mol Biol344:527-47 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Serine protease hepsin |
|---|
| Name: | Serine protease hepsin |
|---|
| Synonyms: | HEPS_HUMAN | HPN | Hepsin | Serine protease hepsin | TMPRSS1 | Transmembrane protease serine 11E | Transmembrane protease, serine 1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 45016.98 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human deglycosylated hepsin was expressed in Pichia pastoris. |
|---|
| Residue: | 417 |
|---|
| Sequence: | MAQKEGGRTVPCCSRPKVAALTAGTLLLLTAIGAASWAIVAVLLRSDQEPLYPVQVSSAD
ARLMVFDKTEGTWRLLCSSRSNARVAGLSCEEMGFLRALTHSELDVRTAGANGTSGFFCV
DEGRLPHTQRLLEVISVCDCPRGRFLAAICQDCGRRKLPVDRIVGGRDTSLGRWPWQVSL
RYDGAHLCGGSLLSGDWVLTAAHCFPERNRVLSRWRVFAGAVAQASPHGLQLGVQAVVYH
GGYLPFRDPNSEENSNDIALVHLSSPLPLTEYIQPVCLPAAGQALVDGKICTVTGWGNTQ
YYGQQAGVLQEARVPIISNDVCNGADFYGNQIKPKMFCAGYPEGGIDACQGDSGGPFVCE
DSISRTPRWRLCGIVSWGTGCALAQKPGVYTKVSDFREWIFQAIKTHSEASGMVTQL
|
|
|
|---|
| BDBM14480 |
|---|
| BDBM13573 |
|---|
| Name | BDBM14480 |
|---|
| Synonyms: | CA-22 | [amino({4-[(3-hydroxynaphthalene-2-)amido]phenyl}amino)methylidene]azanium |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H17N4O2 |
|---|
| Mol. Mass. | 321.3526 |
|---|
| SMILES | [#7]\[#6](-[#7])=[#7+]/c1ccc(-[#7]-[#6](=O)-c2cc3ccccc3cc2-[#8])cc1 |
|---|
| Structure | 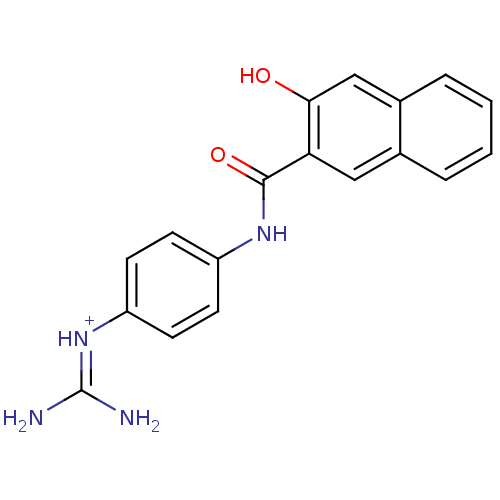
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Katz, BA; Luong, C; Ho, JD; Somoza, JR; Gjerstad, E; Tang, J; Williams, SR; Verner, E; Mackman, RL; Young, WB; Sprengeler, PA; Chan, H; Mortara, K; Janc, JW; McGrath, ME Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA). J Mol Biol344:527-47 (2004) [PubMed] Article
Katz, BA; Luong, C; Ho, JD; Somoza, JR; Gjerstad, E; Tang, J; Williams, SR; Verner, E; Mackman, RL; Young, WB; Sprengeler, PA; Chan, H; Mortara, K; Janc, JW; McGrath, ME Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA). J Mol Biol344:527-47 (2004) [PubMed] Article