| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4B [316-320,321-700,S319G,N320S,N321H,T322M] |
|---|
| Ligand | BDBM14361 |
|---|
| Substrate/Competitor | BDBM10851 |
|---|
| Meas. Tech. | Phosphodiesterase (PDE) Inhibition Assay |
|---|
| IC50 | 570±n/a nM |
|---|
| Citation |  Card, GL; England, BP; Suzuki, Y; Fong, D; Powell, B; Lee, B; Luu, C; Tabrizizad, M; Gillette, S; Ibrahim, PN; Artis, DR; Bollag, G; Milburn, MV; Kim, SH; Schlessinger, J; Zhang, KY Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure12:2233-47 (2004) [PubMed] Article Card, GL; England, BP; Suzuki, Y; Fong, D; Powell, B; Lee, B; Luu, C; Tabrizizad, M; Gillette, S; Ibrahim, PN; Artis, DR; Bollag, G; Milburn, MV; Kim, SH; Schlessinger, J; Zhang, KY Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure12:2233-47 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 4B [316-320,321-700,S319G,N320S,N321H,T322M] |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4B [316-320,321-700,S319G,N320S,N321H,T322M] |
|---|
| Synonyms: | DPDE4 | PDE32 | PDE4B | PDE4B_HUMAN | Phosphodiesterase Type 4 (PDE4B) | cAMP-specific 3,5-cyclic phosphodiesterase 4B |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 45579.64 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | RECOMBINANT CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 4B. |
|---|
| Residue: | 398 |
|---|
| Sequence: | MGSSHHHHHHSSGLVPRGSHMSISRFGVNTENEDHLAKELEDLNKWGLNIFNVAGYSHNR
PLTCIMYAIFQERDLLKTFRISSDTFITYMMTLEDHYHSDVAYHNSLHAADVAQSTHVLL
STPALDAVFTDLEILAAIFAAAIHDVDHPGVSNQFLINTNSELALMYNDESVLENHHLAV
GFKLLQEEHCDIFMNLTKKQRQTLRKMVIDMVLATDMSKHMSLLADLKTMVETKKVTSSG
VLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEISP
MCDKHTASVEKSQVGFIDYIVHPLWETWADLVQPDAQDILDTLEDNRNWYQSMIPQSPSP
PLDEQNRDCQGLMEKFQFELTLDEEDSEGPEKEGEGHS
|
|
|
|---|
| BDBM14361 |
|---|
| BDBM10851 |
|---|
| Name | BDBM14361 |
|---|
| Synonyms: | (R,S)-Rolipram | 4-(3-cyclopentyloxy-4-methoxy-phenyl)pyrrolidin-2-one | 4-[3-(cyclopentyloxy)-4-methoxyphenyl]pyrrolidin-2-one | Adeo | CHEMBL63 | Rolipram |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H21NO3 |
|---|
| Mol. Mass. | 275.3428 |
|---|
| SMILES | COc1ccc(cc1OC1CCCC1)C1CNC(=O)C1 |
|---|
| Structure | 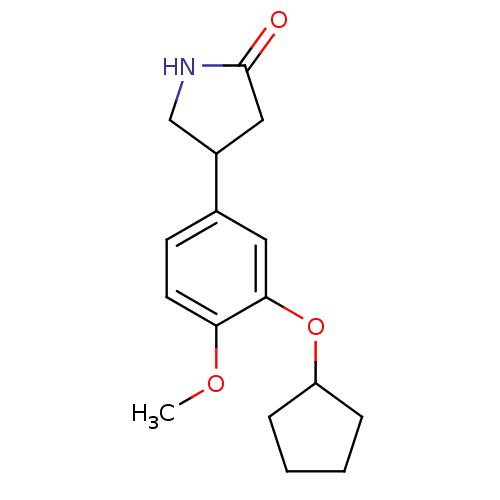
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Card, GL; England, BP; Suzuki, Y; Fong, D; Powell, B; Lee, B; Luu, C; Tabrizizad, M; Gillette, S; Ibrahim, PN; Artis, DR; Bollag, G; Milburn, MV; Kim, SH; Schlessinger, J; Zhang, KY Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure12:2233-47 (2004) [PubMed] Article
Card, GL; England, BP; Suzuki, Y; Fong, D; Powell, B; Lee, B; Luu, C; Tabrizizad, M; Gillette, S; Ibrahim, PN; Artis, DR; Bollag, G; Milburn, MV; Kim, SH; Schlessinger, J; Zhang, KY Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure12:2233-47 (2004) [PubMed] Article