| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Complex of Baculoviral IAP repeat-containing protein 7 [1-159,S150G] and E3 ubiquitin-protein ligase XIAP [336-348] |
|---|
| Ligand | BDBM17343 |
|---|
| Substrate/Competitor | BDBM17342 |
|---|
| Meas. Tech. | Fluorescence Polarization Affinity Measurements. |
|---|
| pH | 7.2±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Ki | 15300±n/a nM |
|---|
| Citation |  Zobel, K; Wang, L; Varfolomeev, E; Franklin, MC; Elliott, LO; Wallweber, HJ; Okawa, DC; Flygare, JA; Vucic, D; Fairbrother, WJ; Deshayes, K Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs. ACS Chem Biol1:525-33 (2006) [PubMed] Article Zobel, K; Wang, L; Varfolomeev, E; Franklin, MC; Elliott, LO; Wallweber, HJ; Okawa, DC; Flygare, JA; Vucic, D; Fairbrother, WJ; Deshayes, K Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs. ACS Chem Biol1:525-33 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Complex of Baculoviral IAP repeat-containing protein 7 [1-159,S150G] and E3 ubiquitin-protein ligase XIAP [336-348] |
|---|
| Name: | Complex of Baculoviral IAP repeat-containing protein 7 [1-159,S150G] and E3 ubiquitin-protein ligase XIAP [336-348] |
|---|
| Synonyms: | Chimeric protein of melanoma inhibitor of apoptosis protein and XIAP-BIR3 | ML-IAP-BIR | MLXBIR3SG |
|---|
| Type: | Chimeric Protein |
|---|
| Mol. Mass.: | 19011.07 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Amino acids 160-179 of MLBIR were replaced with amino acids 336-348 of XIAP-BIR3, and Ser150 of MLBIR was mutated to glycine to give MLXBIR3SG. |
|---|
| Residue: | 172 |
|---|
| Sequence: | MGPKDSAKCLHRGPQPSHWAAGDGPTQERCGPRSLGSPVLGLDTCRAWDHVDGQILGQLR
PLTEEEEEEGAGATLSRGPAFPGMGSEELRLASFYDWPLTAEVPPELLAAAGFFHTGHQD
KVRCFFCYGGLQSWKRGDDPWTEHAKWFPGCQFLLRSKGQEYINNIHLTHSL
|
|
|
|---|
| BDBM17343 |
|---|
| BDBM17342 |
|---|
| Name | BDBM17343 |
|---|
| Synonyms: | (2S)-2-[(2S)-2-[(2S)-6-amino-2-[(2S)-2-[(2S)-2-[(2S,3S)-2-{1-[(3S)-3-[(2S)-2-aminopropanamido]-2-oxoazepan-1-yl]acetamido}-3-methylpentanamido]propanamido]pentanediamido]hexanamido]-3-hydroxypropanamido]pentanedioic acid | Smac-derived compound, 2 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C39H67N11O14 |
|---|
| Mol. Mass. | 914.0146 |
|---|
| SMILES | CCC(C)[C@H](NC(=O)CN1CCCC[C@H](NC(=O)[C@H](C)N)C1=O)C(=O)N[C@@H](C)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CO)C(=O)N[C@@H](CCC(O)=O)C(O)=O |r| |
|---|
| Structure | 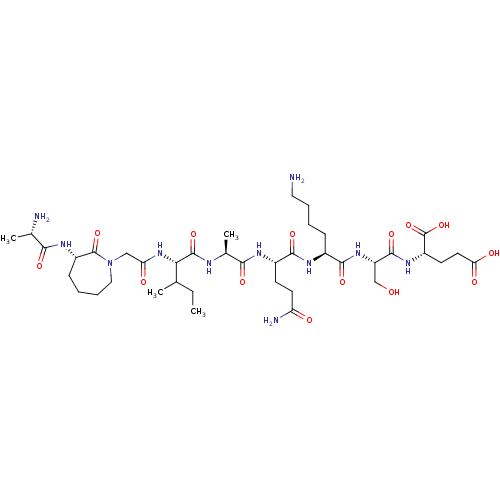
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zobel, K; Wang, L; Varfolomeev, E; Franklin, MC; Elliott, LO; Wallweber, HJ; Okawa, DC; Flygare, JA; Vucic, D; Fairbrother, WJ; Deshayes, K Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs. ACS Chem Biol1:525-33 (2006) [PubMed] Article
Zobel, K; Wang, L; Varfolomeev, E; Franklin, MC; Elliott, LO; Wallweber, HJ; Okawa, DC; Flygare, JA; Vucic, D; Fairbrother, WJ; Deshayes, K Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs. ACS Chem Biol1:525-33 (2006) [PubMed] Article