

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Nociceptin receptor | ||
| Ligand | BDBM21853 | ||
| Substrate/Competitor | BDBM21842 | ||
| Meas. Tech. | Radioligand Binding Assay | ||
| pH | 7.4±n/a | ||
| Temperature | 295.15±n/a K | ||
| Ki | 26±9 nM | ||
| Citation |  Mustazza, C; Borioni, A; Sestili, I; Sbraccia, M; Rodomonte, A; Del Giudice, MR Synthesis and pharmacological evaluation of 1,2-dihydrospiro[isoquinoline-4(3H),4'-piperidin]-3-ones as nociceptin receptor agonists. J Med Chem51:1058-62 (2008) [PubMed] Article Mustazza, C; Borioni, A; Sestili, I; Sbraccia, M; Rodomonte, A; Del Giudice, MR Synthesis and pharmacological evaluation of 1,2-dihydrospiro[isoquinoline-4(3H),4'-piperidin]-3-ones as nociceptin receptor agonists. J Med Chem51:1058-62 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article, Solution Info, Assay Method | ||
| Nociceptin receptor | |||
| Name: | Nociceptin receptor | ||
| Synonyms: | KOR-3 | Kappa-type 3 opioid receptor | Mu-type opioid receptor (Mu) | NOP | Nociceptin Receptor (ORL1 Receptor) | Nociceptin receptor (NOP) | Nociceptin receptor (ORL-1) | Nociceptin receptor (ORL1) | Nociceptin/Orphanin FQ, NOP receptor | OOR | OPIATE ORL-1 | OPRL1 | OPRL1 protein | OPRX_HUMAN | ORL1 | ORL1 receptor | Opioid receptor like-1 | Orphanin FQ receptor | Orphanin FQ receptor (ORL1) | P41146 | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 40702.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P41146 | ||
| Residue: | 370 | ||
| Sequence: |
| ||
| BDBM21853 | |||
| BDBM21842 | |||
| Name | BDBM21853 | ||
| Synonyms: | 2-benzyl-7-fluoro-1'-[4-(propan-2-yl)cyclohexyl]-2,3-dihydro-1H-spiro[isoquinoline-4,4'-piperidine]-3-one | Piperidine Derivative, 5r | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H37FN2O | ||
| Mol. Mass. | 448.6153 | ||
| SMILES | CC(C)C1CCC(CC1)N1CCC2(CC1)C(=O)N(Cc1ccccc1)Cc1cc(F)ccc21 |(.21,-6.77,;-1.12,-6,;-2.46,-6.77,;-1.12,-4.46,;-2.46,-3.69,;-2.46,-2.15,;-1.12,-1.38,;.21,-2.15,;.21,-3.69,;-1.12,.16,;-2.46,.93,;-2.46,2.47,;-1.12,3.24,;.21,2.47,;.21,.93,;.21,4.01,;1.54,3.24,;.21,5.55,;1.54,6.32,;1.54,7.86,;.21,8.63,;.21,10.17,;1.54,10.94,;2.88,10.17,;2.88,8.63,;-1.12,6.32,;-2.46,5.55,;-3.79,6.32,;-5.13,5.55,;-6.46,6.32,;-5.13,4.01,;-3.79,3.24,;-2.46,4.01,)| | ||
| Structure | 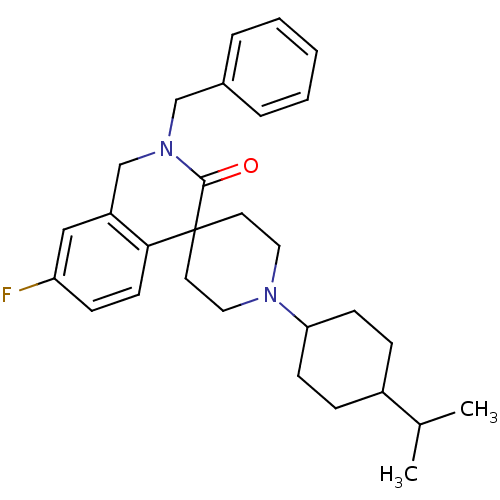
| ||