| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mu-type opioid receptor |
|---|
| Ligand | BDBM21842 |
|---|
| Substrate/Competitor | BDBM21865 |
|---|
| Meas. Tech. | Radioligand Binding Assay |
|---|
| Ki | 3030±n/a nM |
|---|
| Citation |  Mustazza, C; Borioni, A; Sestili, I; Sbraccia, M; Rodomonte, A; Del Giudice, MR Synthesis and pharmacological evaluation of 1,2-dihydrospiro[isoquinoline-4(3H),4'-piperidin]-3-ones as nociceptin receptor agonists. J Med Chem51:1058-62 (2008) [PubMed] Article Mustazza, C; Borioni, A; Sestili, I; Sbraccia, M; Rodomonte, A; Del Giudice, MR Synthesis and pharmacological evaluation of 1,2-dihydrospiro[isoquinoline-4(3H),4'-piperidin]-3-ones as nociceptin receptor agonists. J Med Chem51:1058-62 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Mu-type opioid receptor |
|---|
| Name: | Mu-type opioid receptor |
|---|
| Synonyms: | M-OR-1 | MOP | MOR-1 | MOR1 | MUOR1 | Mu Opioid Receptor | Mu opiate receptor | OPIATE Mu | OPRM1 | OPRM_HUMAN | hMOP | mu-type opioid receptor isoform MOR-1 |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 44789.51 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P35372 |
|---|
| Residue: | 400 |
|---|
| Sequence: | MDSSAAPTNASNCTDALAYSSCSPAPSPGSWVNLSHLDGNLSDPCGPNRTDLGGRDSLCP
PTGSPSMITAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT
STLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDF
RTPRNAKIINVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFI
FAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHI
YVIIKALVTIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCFREFCIPTSSNI
EQQNSTRIRQNTRDHPSTANTVDRTNHQLENLEAETAPLP
|
|
|
|---|
| BDBM21842 |
|---|
| BDBM21865 |
|---|
| Name | BDBM21842 |
|---|
| Synonyms: | (2S)-2-[(2S)-2-[(2S)-2-[(2S)-2-[(2S)-6-amino-2-[(2S)-2-[(2S)-2-[(2S)-2-[(2S)-6-amino-2-[(2S)-2-[(2S)-2-{2-[(2S,3R)-2-[(2S)-2-(2-{2-[(2S)-2-amino-3-phenylpropanamido]acetamido}acetamido)-3-phenylpropanamido]-3-hydroxybutanamido]acetamido}propanamido]-5-carbamimidamidopentanamido]hexanamido]-3-hydroxypropanamido]propanamido]-5-carbamimidamidopentanamido]hexanamido]-4-methylpentanamido]propanamido]butanediamido]-4-carbamoylbutanoic acid | N/OFQ | OFQ 1-17 | OFQ/N | Orphanin FQ | [125I]-nociceptin | nociceptin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C79H129N27O22 |
|---|
| Mol. Mass. | 1809.0373 |
|---|
| SMILES | [#6]-[#6](-[#6])-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#7]-[#6](=O)-[#6@H](-[#6])-[#7]-[#6](=O)-[#6@H](-[#6]-[#8])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#7]-[#6](=O)-[#6@H](-[#6])-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@@H](-[#7])-[#6]-c1ccccc1)-[#6@@H](-[#6])-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#6](-[#8])=O |
|---|
| Structure | 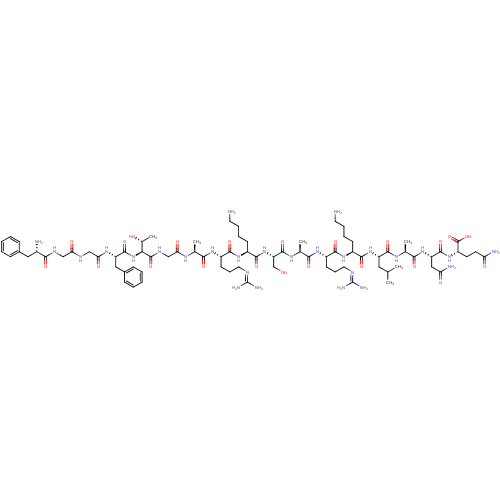
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mustazza, C; Borioni, A; Sestili, I; Sbraccia, M; Rodomonte, A; Del Giudice, MR Synthesis and pharmacological evaluation of 1,2-dihydrospiro[isoquinoline-4(3H),4'-piperidin]-3-ones as nociceptin receptor agonists. J Med Chem51:1058-62 (2008) [PubMed] Article
Mustazza, C; Borioni, A; Sestili, I; Sbraccia, M; Rodomonte, A; Del Giudice, MR Synthesis and pharmacological evaluation of 1,2-dihydrospiro[isoquinoline-4(3H),4'-piperidin]-3-ones as nociceptin receptor agonists. J Med Chem51:1058-62 (2008) [PubMed] Article