| Reaction Details |
|---|
 | Report a problem with these data |
| Target | S-methyl-5'-thioadenosine phosphorylase |
|---|
| Ligand | BDBM22113 |
|---|
| Substrate/Competitor | BDBM22111 |
|---|
| Meas. Tech. | MTAP/MTAN Inhibition Assay |
|---|
| pH | 7±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Ki | 1.7±0.2 nM |
|---|
| Km | 5000±n/a nM |
|---|
| Comments | Ki is the dissociation constant for the first step in the two-step binding characteristic of slow-onset tight-binding inhibition. |
|---|
| Citation |  Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| S-methyl-5'-thioadenosine phosphorylase |
|---|
| Name: | S-methyl-5'-thioadenosine phosphorylase |
|---|
| Synonyms: | 5'-Methylthioadenosine phosphorylase (MTAP) | MSAP | MTA phosphorylase | MTAP | MTAP_HUMAN | MTAPase | Methylthioadenosine Phosphorylase (MTAP) | S-methyl-5 -thioadenosine phosphorylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 31239.23 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 283 |
|---|
| Sequence: | MASGTTTTAVKIGIIGGTGLDDPEILEGRTEKYVDTPFGKPSDALILGKIKNVDCVLLAR
HGRQHTIMPSKVNYQANIWALKEEGCTHVIVTTACGSLREEIQPGDIVIIDQFIDRTTMR
PQSFYDGSHSCARGVCHIPMAEPFCPKTREVLIETAKKLGLRCHSKGTMVTIEGPRFSSR
AESFMFRTWGADVINMTTVPEVVLAKEAGICYASIAMATDYDCWKEHEEAVSVDRVLKTL
KENANKAKSLLLTTIPQIGSTEWSETLHNLKNMAQFSVLLPRH
|
|
|
|---|
| BDBM22113 |
|---|
| BDBM22111 |
|---|
| Name | BDBM22113 |
|---|
| Synonyms: | (3R,4S)-1-({4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl}methyl)-4-[(methylsulfanyl)methyl]pyrrolidin-3-ol | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(methylsulfanyl)methyl]pyrrolidin-3-ol | (3R,4S)-1-[(9-Deazaadenin-9-yl)methyl]-3-hydroxy-4-methylthiomethylpyrrolidine, 7 | DADMe-ImmA-Me | MT-DADMe-ImmA |
|---|
| Type | n/a |
|---|
| Emp. Form. | C13H19N5OS |
|---|
| Mol. Mass. | 293.388 |
|---|
| SMILES | CSC[C@H]1CN(Cc2c[nH]c3c(N)ncnc23)C[C@@H]1O |r| |
|---|
| Structure | 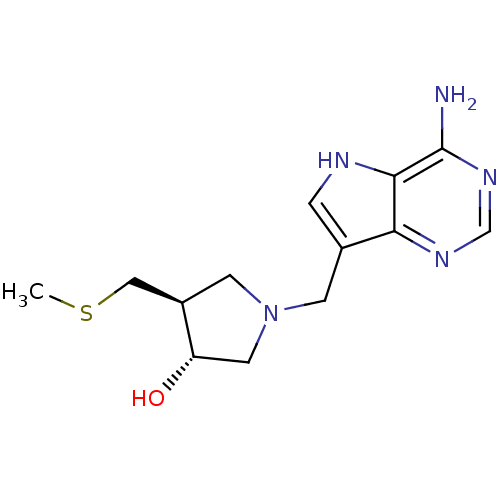
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article
Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article