| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Sodium-dependent dopamine transporter |
|---|
| Ligand | BDBM22199 |
|---|
| Substrate/Competitor | BDBM22166 |
|---|
| Meas. Tech. | Radioligand Binding Assay |
|---|
| Ki | 0.9±n/a nM |
|---|
| Citation |  Hsin, LW; Chang, LT; Rothman, RB; Dersch, CM; Jacobson, AE; Rice, KC Design and Synthesis of 2- and 3-Substituted-3-phenylpropyl Analogs of 1-[2-[Bis(4-fluorophenyl)methoxy]ethyl]-4-(3-phenylpropyl)piperazine and 1-[2-(Diphenylmethoxy)ethyl]-4-(3-phenylpropyl)piperazine: Role of Amino, Fluoro, Hydroxyl, Methoxyl, Methyl, Methylene, and Oxo Substituents on Affinity f J Med Chem51:2795-806 (2008) [PubMed] Article Hsin, LW; Chang, LT; Rothman, RB; Dersch, CM; Jacobson, AE; Rice, KC Design and Synthesis of 2- and 3-Substituted-3-phenylpropyl Analogs of 1-[2-[Bis(4-fluorophenyl)methoxy]ethyl]-4-(3-phenylpropyl)piperazine and 1-[2-(Diphenylmethoxy)ethyl]-4-(3-phenylpropyl)piperazine: Role of Amino, Fluoro, Hydroxyl, Methoxyl, Methyl, Methylene, and Oxo Substituents on Affinity f J Med Chem51:2795-806 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Sodium-dependent dopamine transporter |
|---|
| Name: | Sodium-dependent dopamine transporter |
|---|
| Synonyms: | DA transporter | Monoamine transporters; Norepininephrine & dopamine | SC6A3_RAT | Slc6a3 | Sodium-dependent dopamine transporter | Sodium-dependent dopamine transporter (DAT) |
|---|
| Type: | Multi-pass membrane protein |
|---|
| Mol. Mass.: | 68749.45 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | P23977 |
|---|
| Residue: | 619 |
|---|
| Sequence: | MSKSKCSVGPMSSVVAPAKESNAVGPREVELILVKEQNGVQLTNSTLINPPQTPVEAQER
ETWSKKIDFLLSVIGFAVDLANVWRFPYLCYKNGGGAFLVPYLLFMVIAGMPLFYMELAL
GQFNREGAAGVWKICPVLKGVGFTVILISFYVGFFYNVIIAWALHYFFSSFTMDLPWIHC
NNTWNSPNCSDAHASNSSDGLGLNDTFGTTPAAEYFERGVLHLHQSRGIDDLGPPRWQLT
ACLVLVIVLLYFSLWKGVKTSGKVVWITATMPYVVLTALLLRGVTLPGAMDGIRAYLSVD
FYRLCEASVWIDAATQVCFSLGVGFGVLIAFSSYNKFTNNCYRDAIITTSINSLTSFSSG
FVVFSFLGYMAQKHNVPIRDVATDGPGLIFIIYPEAIATLPLSSAWAAVFFLMLLTLGID
SAMGGMESVITGLVDEFQLLHRHRELFTLGIVLATFLLSLFCVTNGGIYVFTLLDHFAAG
TSILFGVLIEAIGVAWFYGVQQFSDDIKQMTGQRPNLYWRLCWKLVSPCFLLYVVVVSIV
TFRPPHYGAYIFPDWANALGWIIATSSMAMVPIYATYKFCSLPGSFREKLAYAITPEKDH
QLVDRGEVRQFTLRHWLLL
|
|
|
|---|
| BDBM22199 |
|---|
| BDBM22166 |
|---|
| Name | BDBM22199 |
|---|
| Synonyms: | 1-{2-[bis(4-fluorophenyl)methoxy]ethyl}-4-[(2E)-3-phenylprop-2-en-1-yl]piperazine | CHEMBL112280 | CHEMBL286991 | GBR 12935 Analogue, 36 | GBR-13,069 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H30F2N2O |
|---|
| Mol. Mass. | 448.5474 |
|---|
| SMILES | Fc1ccc(cc1)C(OCCN1CCN(C\C=C\c2ccccc2)CC1)c1ccc(F)cc1 |
|---|
| Structure | 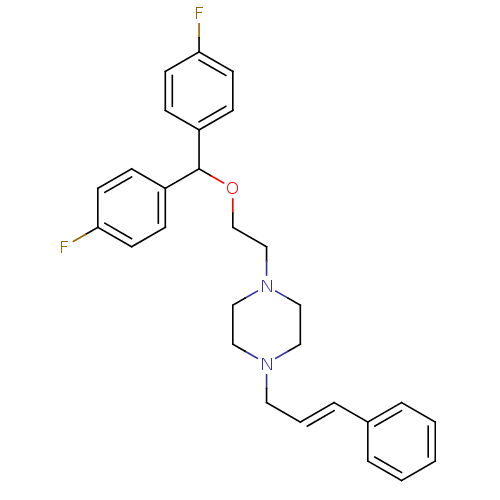
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hsin, LW; Chang, LT; Rothman, RB; Dersch, CM; Jacobson, AE; Rice, KC Design and Synthesis of 2- and 3-Substituted-3-phenylpropyl Analogs of 1-[2-[Bis(4-fluorophenyl)methoxy]ethyl]-4-(3-phenylpropyl)piperazine and 1-[2-(Diphenylmethoxy)ethyl]-4-(3-phenylpropyl)piperazine: Role of Amino, Fluoro, Hydroxyl, Methoxyl, Methyl, Methylene, and Oxo Substituents on Affinity f J Med Chem51:2795-806 (2008) [PubMed] Article
Hsin, LW; Chang, LT; Rothman, RB; Dersch, CM; Jacobson, AE; Rice, KC Design and Synthesis of 2- and 3-Substituted-3-phenylpropyl Analogs of 1-[2-[Bis(4-fluorophenyl)methoxy]ethyl]-4-(3-phenylpropyl)piperazine and 1-[2-(Diphenylmethoxy)ethyl]-4-(3-phenylpropyl)piperazine: Role of Amino, Fluoro, Hydroxyl, Methoxyl, Methyl, Methylene, and Oxo Substituents on Affinity f J Med Chem51:2795-806 (2008) [PubMed] Article