| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Calpain-1 catalytic subunit |
|---|
| Ligand | BDBM23863 |
|---|
| Substrate/Competitor | BDBM23856 |
|---|
| Meas. Tech. | mu-Calpain Inhibition Assay |
|---|
| pH | 7.4±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Ki | 63±4 nM |
|---|
| Citation |  Donkor, IO; Assefa, H; Liu, J Structural basis for the potent calpain inhibitory activity of peptidyl alpha-ketoacids. J Med Chem51:4346-50 (2008) [PubMed] Article Donkor, IO; Assefa, H; Liu, J Structural basis for the potent calpain inhibitory activity of peptidyl alpha-ketoacids. J Med Chem51:4346-50 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Calpain-1 catalytic subunit |
|---|
| Name: | Calpain-1 catalytic subunit |
|---|
| Synonyms: | CAN1_PIG | CANP 1 | CAPN1 | Calcium-activated neutral proteinase 1 | Calpain 1 | Calpain mu-type | Calpain-1 | Calpain-1 catalytic subunit | Calpain-1 large subunit | Micromolar-calpain | muCANP |
|---|
| Type: | Catalytic subunit; forms a heterodimer with a small (regulatory) subunit (CAPNS1) |
|---|
| Mol. Mass.: | 81729.82 |
|---|
| Organism: | Sus scrofa (pig) |
|---|
| Description: | Native calpain-1 from porcine erythrocytes, purchased from Calbiochem, was used in assay. |
|---|
| Residue: | 714 |
|---|
| Sequence: | MAEEVITPVYCTGVSAQVQKLRAKELGLGRHENAIKYLGQDYEQLRAHCLQSGSLFRDEA
FPPVPQSLGFKELGPNSSKTYGVKWKRPTELFSNPQFIVDGATRTDICQGALGDCWLLAA
IASLTLNDTLLHRVVPHGQSFQNGYAGIFHFQLWQFGEWVDVVVDDLLPTKDGKLVFVHS
AQGNEFWSALLEKAYAKVNGSYEALSGGSTSEGFEDFTGGVTEWYELRKAPSDLYSIILK
ALERGSLLGCSIDISSVLDMEAVTFKKLVKGHAYSVTGAKQVNYQGQMVNLIRMRNPWGE
VEWTGAWSDGSSEWNGVDPYQRDQLRVRMEDGEFWMSFRDFLREFTRLEICNLTPDALKS
QRVRNWNTTLYEGTWRRGSTAGGCRNYPATFWVNPQFKIRLEETDDPEDDYGGRESGCSF
VLALMQKHRRRERRFGRDMETIGFAVYEVPPELVGQPVHLKRDFFLANASRARSEQFINL
REVSTRFRLPPGEYVVVPSTFEPNKEGDFVLRFFSEKKAGTQELDDQVQAILPDEQVLSE
EEIDENFKALFRQLAGEDMEISVRELRTILNRIISKHKDLRTKGFSLESCRSMVNLMDRD
GNGKLGLVEFNILWNRIRNYLSIFRKFDLDKSGSMSAYEMRMAIESAGFKLNKKLFELII
TRYSEPDLAVDFDNFVCCLVRLETMFRFFKTLDTDLDGVVTFDLFKWLQLTMFA
|
|
|
|---|
| BDBM23863 |
|---|
| BDBM23856 |
|---|
| Name | BDBM23863 |
|---|
| Synonyms: | 3-[(2S)-2-(3,3-diphenylpropanamido)-4-methylpentanamido]-2-oxo-4-phenylbutanoic acid | Peptidyl alpha-Ketoacid, 2d |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H34N2O5 |
|---|
| Mol. Mass. | 514.6121 |
|---|
| SMILES | CC(C)C[C@H](NC(=O)CC(c1ccccc1)c1ccccc1)C(=O)NC(Cc1ccccc1)C(=O)C(O)=O |r| |
|---|
| Structure | 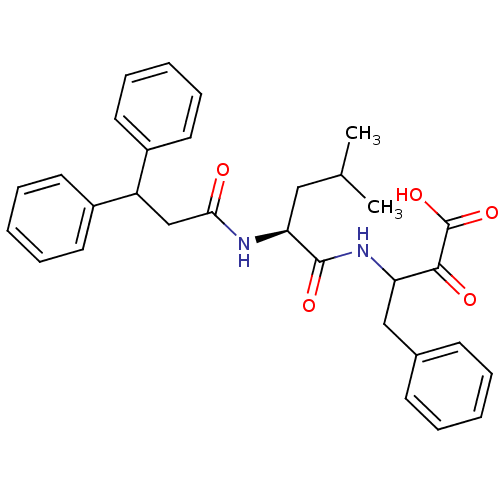
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Donkor, IO; Assefa, H; Liu, J Structural basis for the potent calpain inhibitory activity of peptidyl alpha-ketoacids. J Med Chem51:4346-50 (2008) [PubMed] Article
Donkor, IO; Assefa, H; Liu, J Structural basis for the potent calpain inhibitory activity of peptidyl alpha-ketoacids. J Med Chem51:4346-50 (2008) [PubMed] Article