| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 4 [653-1084,H976Y] |
|---|
| Ligand | BDBM25150 |
|---|
| Substrate/Competitor | BDBM25143 |
|---|
| Meas. Tech. | HDAC Activity Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 310.15±n/a K |
|---|
| IC50 | 24±n/a nM |
|---|
| Citation |  Jones, P; Altamura, S; De Francesco, R; Gallinari, P; Lahm, A; Neddermann, P; Rowley, M; Serafini, S; Steinkühler, C Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett18:1814-9 (2008) [PubMed] Article Jones, P; Altamura, S; De Francesco, R; Gallinari, P; Lahm, A; Neddermann, P; Rowley, M; Serafini, S; Steinkühler, C Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett18:1814-9 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Histone deacetylase 4 [653-1084,H976Y] |
|---|
| Name: | Histone deacetylase 4 [653-1084,H976Y] |
|---|
| Synonyms: | HDAC4 | HDAC4_HUMAN | Histone Deacetylase 4 (HDAC4) Mutant (H976Y) | Histone deacetylase 4 gain of function (GOF) mutant | KIAA0288 |
|---|
| Type: | Catalytic domain |
|---|
| Mol. Mass.: | 46476.72 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | His-tagged HDAC4 GOF(gain of function, H976Y) catalytic domains (T653-L1084) was expressed in E.coli and purified by Nickel-Chelation affinity chromatography and anion exchange (MonoQ) chromatography. |
|---|
| Residue: | 432 |
|---|
| Sequence: | TTGLVYDTLMLKHQCTCGSSSSHPEHAGRIQSIWSRLQETGLRGKCECIRGRKATLEELQ
TVHSEAHTLLYGTNPLNRQKLDSKKLLGSLASVFVRLPCGGVGVDSDTIWNEVHSAGAAR
LAVGCVVELVFKVATGELKNGFAVVRPPGHHAEESTPMGFCYFNSVAVAAKLLQQRLSVS
KILIVDWDVHHGNGTQQAFYSDPSVLYMSLHRYDDGNFFPGSGAPDEVGTGPGVGFNVNM
AFTGGLDPPMGDAEYLAAFRTVVMPIASEFAPDVVLVSSGFDAVEGHPTPLGGYNLSARC
FGYLTKQLMGLAGGRIVLALEGGYDLTAICDASEACVSALLGNELDPLPEKVLQQRPNAN
AVRSMEKVMEIHSKYWRCLQRTTSTAGRSLIEAQTCENEEAETVTAMASLSVGVKPAEKR
PDEEPMEEEPPL
|
|
|
|---|
| BDBM25150 |
|---|
| BDBM25143 |
|---|
| Name | BDBM25150 |
|---|
| Synonyms: | (2E)-N-hydroxy-3-[3-(phenylsulfamoyl)phenyl]prop-2-enamide | Belinosta | Belinostat | PXD-101 | PXD101 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H14N2O4S |
|---|
| Mol. Mass. | 318.348 |
|---|
| SMILES | ONC(=O)\C=C\c1cccc(c1)S(=O)(=O)Nc1ccccc1 |
|---|
| Structure | 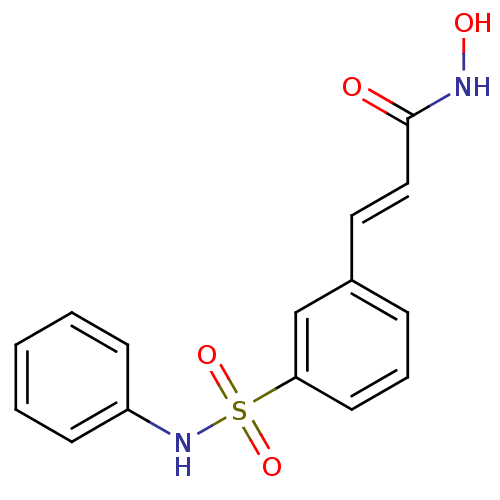
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jones, P; Altamura, S; De Francesco, R; Gallinari, P; Lahm, A; Neddermann, P; Rowley, M; Serafini, S; Steinkühler, C Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett18:1814-9 (2008) [PubMed] Article
Jones, P; Altamura, S; De Francesco, R; Gallinari, P; Lahm, A; Neddermann, P; Rowley, M; Serafini, S; Steinkühler, C Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett18:1814-9 (2008) [PubMed] Article