| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Adenosine 5'-phosphosulfate reductase |
|---|
| Ligand | BDBM25465 |
|---|
| Substrate/Competitor | BDBM25461 |
|---|
| Meas. Tech. | Determination of Dissociation Constant (Kd) |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 303.15±n/a K |
|---|
| Kd | 127190±n/a nM |
|---|
| Citation |  Cosconati, S; Hong, JA; Novellino, E; Carroll, KS; Goodsell, DS; Olson, AJ Structure-based virtual screening and biological evaluation of Mycobacterium tuberculosis adenosine 5'-phosphosulfate reductase inhibitors. J Med Chem51:6627-30 (2008) [PubMed] Article Cosconati, S; Hong, JA; Novellino, E; Carroll, KS; Goodsell, DS; Olson, AJ Structure-based virtual screening and biological evaluation of Mycobacterium tuberculosis adenosine 5'-phosphosulfate reductase inhibitors. J Med Chem51:6627-30 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Adenosine 5'-phosphosulfate reductase |
|---|
| Name: | Adenosine 5'-phosphosulfate reductase |
|---|
| Synonyms: | 3-phosphoadenylylsulfate reductase | Adenosine Phosphosulfate Reductase (APSR) | CYSH_MYCTO | PAPS reductase, thioredoxin dependent | PAPS sulfotransferase | PAdoPS reductase | cysH | phosphoadenosine phosphosulfate reductase |
|---|
| Type: | Oxidoreductase |
|---|
| Mol. Mass.: | 27417.08 |
|---|
| Organism: | Mycobacterium tuberculosis |
|---|
| Description: | Proteins were expressed by transforming a reductase-containing plasmid into E. coli cells. |
|---|
| Residue: | 254 |
|---|
| Sequence: | MSGETTRLTEPQLRELAARGAAELDGATATDMLRWTDETFGDIGGAGGGVSGHRGWTTCN
YVVASNMADAVLVDLAAKVRPGVPVIFLDTGYHFVETIGTRDAIESVYDVRVLNVTPEHT
VAEQDELLGKDLFARNPHECCRLRKVVPLGKTLRGYSAWVTGLRRVDAPTRANAPLVSFD
ETFKLVKVNPLAAWTDQDVQEYIADNDVLVNPLVREGYPSIGCAPCTAKPAEGADPRSGR
WQGLAKTECGLHAS
|
|
|
|---|
| BDBM25465 |
|---|
| BDBM25461 |
|---|
| Name | BDBM25465 |
|---|
| Synonyms: | (1R,2R,5S,6S,10S,13S,15R)-5-acetyl-13-hydroxy-6,10-dimethylpentacyclo[13.2.2.0^{1,9}.0^{2,6}.0^{10,15}]nonadeca-8,18-diene-16,17-dicarboxylic acid | NSC9746 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H32O6 |
|---|
| Mol. Mass. | 428.518 |
|---|
| SMILES | [H][C@@]12CC[C@H](C(C)=O)[C@@]1(C)CC=C1[C@@]3(C)CC[C@H](O)C[C@]33C=C[C@]21C(C3C(O)=O)C(O)=O |c:23,t:12,THB:15:13:24.25:22.21,11:12:24.25:22.21,14:13:24.25:22.21| |
|---|
| Structure | 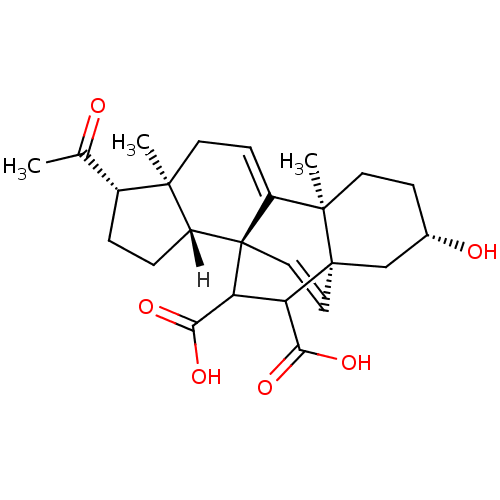
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cosconati, S; Hong, JA; Novellino, E; Carroll, KS; Goodsell, DS; Olson, AJ Structure-based virtual screening and biological evaluation of Mycobacterium tuberculosis adenosine 5'-phosphosulfate reductase inhibitors. J Med Chem51:6627-30 (2008) [PubMed] Article
Cosconati, S; Hong, JA; Novellino, E; Carroll, KS; Goodsell, DS; Olson, AJ Structure-based virtual screening and biological evaluation of Mycobacterium tuberculosis adenosine 5'-phosphosulfate reductase inhibitors. J Med Chem51:6627-30 (2008) [PubMed] Article