| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Carbonic anhydrase 13 |
|---|
| Ligand | BDBM26998 |
|---|
| Substrate/Competitor | BDBM10856 |
|---|
| Meas. Tech. | CA Inhibition Assay |
|---|
| Ki | 645±n/a nM |
|---|
| Citation |  Temperini, C; Cecchi, A; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors. Comparison of chlorthalidone, indapamide, trichloromethiazide, and furosemide X-ray crystal structures in adducts with isozyme II, when several water molecules make the difference. Bioorg Med Chem17:1214-21 (2009) [PubMed] Article Temperini, C; Cecchi, A; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors. Comparison of chlorthalidone, indapamide, trichloromethiazide, and furosemide X-ray crystal structures in adducts with isozyme II, when several water molecules make the difference. Bioorg Med Chem17:1214-21 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Carbonic anhydrase 13 |
|---|
| Name: | Carbonic anhydrase 13 |
|---|
| Synonyms: | CA-XIII | CAH13_MOUSE | Ca13 | Car13 | Carbonate dehydratase XIII | Carbonic anhydrase 13 | Carbonic anhydrase XIII |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 29522.80 |
|---|
| Organism: | Mus musculus (mouse) |
|---|
| Description: | Murine cloned isozyme |
|---|
| Residue: | 262 |
|---|
| Sequence: | MARLSWGYGEHNGPIHWNELFPIADGDQQSPIEIKTKEVKYDSSLRPLSIKYDPASAKII
SNSGHSFNVDFDDTEDKSVLRGGPLTGNYRLRQFHLHWGSADDHGSEHVVDGVRYAAELH
VVHWNSDKYPSFVEAAHESDGLAVLGVFLQIGEHNPQLQKITDILDSIKEKGKQTRFTNF
DPLCLLPSSWDYWTYPGSLTVPPLLESVTWIVLKQPISISSQQLARFRSLLCTAEGESAA
FLLSNHRPPQPLKGRRVRASFY
|
|
|
|---|
| BDBM26998 |
|---|
| BDBM10856 |
|---|
| Name | BDBM26998 |
|---|
| Synonyms: | 6-chloro-3-(dichloromethyl)-1,1-dioxo-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide | TRICHLORMETHIAZIDE | trichloromethiazide, 6 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C8H8Cl3N3O4S2 |
|---|
| Mol. Mass. | 380.656 |
|---|
| SMILES | NS(=O)(=O)c1cc2c(NC(NS2(=O)=O)C(Cl)Cl)cc1Cl |
|---|
| Structure | 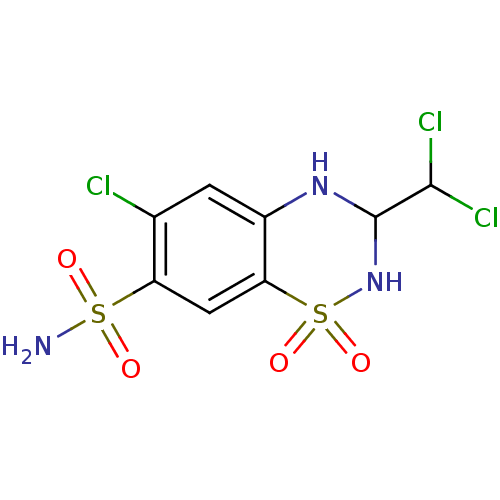
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Temperini, C; Cecchi, A; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors. Comparison of chlorthalidone, indapamide, trichloromethiazide, and furosemide X-ray crystal structures in adducts with isozyme II, when several water molecules make the difference. Bioorg Med Chem17:1214-21 (2009) [PubMed] Article
Temperini, C; Cecchi, A; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors. Comparison of chlorthalidone, indapamide, trichloromethiazide, and furosemide X-ray crystal structures in adducts with isozyme II, when several water molecules make the difference. Bioorg Med Chem17:1214-21 (2009) [PubMed] Article