| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Ligand | BDBM27233 |
|---|
| Substrate/Competitor | BDBM27221 |
|---|
| Meas. Tech. | Fluorescence Polarization-Based Binding Displacement Kinase Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 303.15±n/a K |
|---|
| Kd | 443±11 nM |
|---|
| Citation |  Lavogina, D; Lust, M; Viil, I; König, N; Raidaru, G; Rogozina, J; Enkvist, E; Uri, A; Bossemeyer, D Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases. J Med Chem52:308-21 (2009) [PubMed] Article Lavogina, D; Lust, M; Viil, I; König, N; Raidaru, G; Rogozina, J; Enkvist, E; Uri, A; Bossemeyer, D Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases. J Med Chem52:308-21 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Name: | cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Synonyms: | KAPCA_MOUSE | PKA C-alpha | Pkaca | Prkaca | cAMP-Dependent Protein Kinase (PKA) | cAMP-dependent protein kinase catalytic subunit alpha | cAMP-dependent protein kinase, alpha-catalytic subunit |
|---|
| Type: | Enzyme Catalytic Unit |
|---|
| Mol. Mass.: | 40579.18 |
|---|
| Organism: | Mus musculus (mouse) |
|---|
| Description: | n/a |
|---|
| Residue: | 351 |
|---|
| Sequence: | MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVML
VKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV
MEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY
IQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF
ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT
TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFTEF
|
|
|
|---|
| BDBM27233 |
|---|
| BDBM27221 |
|---|
| Name | BDBM27233 |
|---|
| Synonyms: | (2R)-6-amino-2-(6-{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]formamido}hexanamido)hexanamide | ARC-1030, 7 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H35N9O6 |
|---|
| Mol. Mass. | 521.57 |
|---|
| SMILES | NCCCC[C@@H](NC(=O)CCCCCNC(=O)[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(N)ncnc12)C(N)=O |r| |
|---|
| Structure | 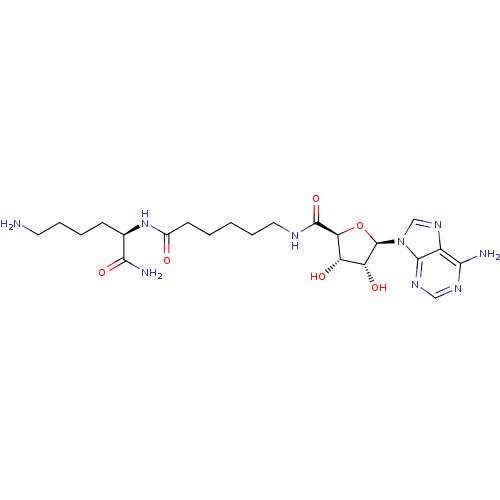
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lavogina, D; Lust, M; Viil, I; König, N; Raidaru, G; Rogozina, J; Enkvist, E; Uri, A; Bossemeyer, D Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases. J Med Chem52:308-21 (2009) [PubMed] Article
Lavogina, D; Lust, M; Viil, I; König, N; Raidaru, G; Rogozina, J; Enkvist, E; Uri, A; Bossemeyer, D Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases. J Med Chem52:308-21 (2009) [PubMed] Article