| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 1 |
|---|
| Ligand | BDBM27770 |
|---|
| Substrate/Competitor | BDBM25143 |
|---|
| Meas. Tech. | HDAC Activity Assay |
|---|
| pH | 8±n/a |
|---|
| Temperature | 310.15±n/a K |
|---|
| IC50 | 10±n/a nM |
|---|
| Citation |  Kattar, SD; Surdi, LM; Zabierek, A; Methot, JL; Middleton, RE; Hughes, B; Szewczak, AA; Dahlberg, WK; Kral, AM; Ozerova, N; Fleming, JC; Wang, H; Secrist, P; Harsch, A; Hamill, JE; Cruz, JC; Kenific, CM; Chenard, M; Miller, TA; Berk, SC; Tempest, P Parallel medicinal chemistry approaches to selective HDAC1/HDAC2 inhibitor (SHI-1:2) optimization. Bioorg Med Chem Lett19:1168-72 (2009) [PubMed] Article Kattar, SD; Surdi, LM; Zabierek, A; Methot, JL; Middleton, RE; Hughes, B; Szewczak, AA; Dahlberg, WK; Kral, AM; Ozerova, N; Fleming, JC; Wang, H; Secrist, P; Harsch, A; Hamill, JE; Cruz, JC; Kenific, CM; Chenard, M; Miller, TA; Berk, SC; Tempest, P Parallel medicinal chemistry approaches to selective HDAC1/HDAC2 inhibitor (SHI-1:2) optimization. Bioorg Med Chem Lett19:1168-72 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Histone deacetylase 1 |
|---|
| Name: | Histone deacetylase 1 |
|---|
| Synonyms: | Cereblon/Histone deacetylase 1 | HD1 | HDAC1 | HDAC1_HUMAN | Histone deacetylase 1 (HDAC1) | Human HDAC1 | RPD3L1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55090.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13547 |
|---|
| Residue: | 482 |
|---|
| Sequence: | MAQTQGTRRKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKAN
AEEMTKYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGEDCPVFDGLFEFCQLSTGGSVAS
AVKLNKQQTDIAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHG
DGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAI
FKPVMSKVMEMFQPSAVVLQCGSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNLPMLMLGG
GGYTIRNVARCWTYETAVALDTEIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTNEYLE
KIKQRLFENLRMLPHAPGVQMQAIPEDAIPEESGDEDEDDPDKRISICSSDKRIACEEEF
SDSEEEGEGGRKNSSNFKKAKRVKTEDEKEKDPEEKKEVTEEEKTKEEKPEAKGVKEEVK
LA
|
|
|
|---|
| BDBM27770 |
|---|
| BDBM25143 |
|---|
| Name | BDBM27770 |
|---|
| Synonyms: | N-(2-amino-5-phenylphenyl)-4-{[methyl(pyridin-4-yl)amino]methyl}benzamide | US9096559, 62 | biphenyl diamine analogue, 18 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H24N4O |
|---|
| Mol. Mass. | 408.495 |
|---|
| SMILES | CN(Cc1ccc(cc1)C(=O)Nc1cc(ccc1N)-c1ccccc1)c1ccncc1 |
|---|
| Structure | 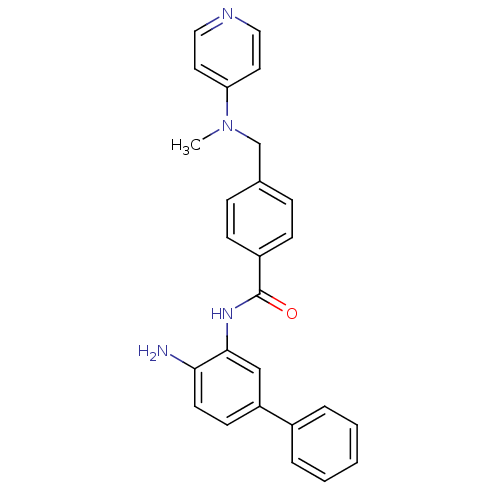
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kattar, SD; Surdi, LM; Zabierek, A; Methot, JL; Middleton, RE; Hughes, B; Szewczak, AA; Dahlberg, WK; Kral, AM; Ozerova, N; Fleming, JC; Wang, H; Secrist, P; Harsch, A; Hamill, JE; Cruz, JC; Kenific, CM; Chenard, M; Miller, TA; Berk, SC; Tempest, P Parallel medicinal chemistry approaches to selective HDAC1/HDAC2 inhibitor (SHI-1:2) optimization. Bioorg Med Chem Lett19:1168-72 (2009) [PubMed] Article
Kattar, SD; Surdi, LM; Zabierek, A; Methot, JL; Middleton, RE; Hughes, B; Szewczak, AA; Dahlberg, WK; Kral, AM; Ozerova, N; Fleming, JC; Wang, H; Secrist, P; Harsch, A; Hamill, JE; Cruz, JC; Kenific, CM; Chenard, M; Miller, TA; Berk, SC; Tempest, P Parallel medicinal chemistry approaches to selective HDAC1/HDAC2 inhibitor (SHI-1:2) optimization. Bioorg Med Chem Lett19:1168-72 (2009) [PubMed] Article