| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Uridine 5'-monophosphate synthase |
|---|
| Ligand | BDBM27943 |
|---|
| Substrate/Competitor | BDBM21336 |
|---|
| Meas. Tech. | Enzyme Inhibition Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 310.15±n/a K |
|---|
| Ki | 98000±6000 nM |
|---|
| Citation |  Bello, AM; Konforte, D; Poduch, E; Furlonger, C; Wei, L; Liu, Y; Lewis, M; Pai, EF; Paige, CJ; Kotra, LP Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents. J Med Chem52:1648-58 (2009) [PubMed] Article Bello, AM; Konforte, D; Poduch, E; Furlonger, C; Wei, L; Liu, Y; Lewis, M; Pai, EF; Paige, CJ; Kotra, LP Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents. J Med Chem52:1648-58 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Uridine 5'-monophosphate synthase |
|---|
| Name: | Uridine 5'-monophosphate synthase |
|---|
| Synonyms: | Orotate phosphoribosyltransferase (HsOPRT) | Orotidine Monophosphate Decarboxylase (ODCase) | UMPS | UMPS_HUMAN |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52224.99 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P11172 |
|---|
| Residue: | 480 |
|---|
| Sequence: | MAVARAALGPLVTGLYDVQAFKFGDFVLKSGLSSPIYIDLRGIVSRPRLLSQVADILFQT
AQNAGISFDTVCGVPYTALPLATVICSTNQIPMLIRRKETKDYGTKRLVEGTINPGETCL
IIEDVVTSGSSVLETVEVLQKEGLKVTDAIVLLDREQGGKDKLQAHGIRLHSVCTLSKML
EILEQQKKVDAETVGRVKRFIQENVFVAANHNGSPLSIKEAPKELSFGARAELPRIHPVA
SKLLRLMQKKETNLCLSADVSLARELLQLADALGPSICMLKTHVDILNDFTLDVMKELIT
LAKCHEFLIFEDRKFADIGNTVKKQYEGGIFKIASWADLVNAHVVPGSGVVKGLQEVGLP
LHRGCLLIAEMSSTGSLATGDYTRAAVRMAEEHSEFVVGFISGSRVSMKPEFLHLTPGVQ
LEAGGDNLGQQYNSPQEVIGKRGSDIIIVGRGIISAADRLEAAEMYRKAAWEAYLSRLGV
|
|
|
|---|
| BDBM27943 |
|---|
| BDBM21336 |
|---|
| Name | BDBM27943 |
|---|
| Synonyms: | uridine derivative, 39 | {[(2R,3S,4R,5R)-5-(5-fluoro-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid |
|---|
| Type | n/a |
|---|
| Emp. Form. | C9H12FN2O9P |
|---|
| Mol. Mass. | 342.1717 |
|---|
| SMILES | O[C@H]1[C@@H](O)[C@@H](O[C@@H]1COP(O)(O)=O)n1cc(F)c(=O)[nH]c1=O |r| |
|---|
| Structure | 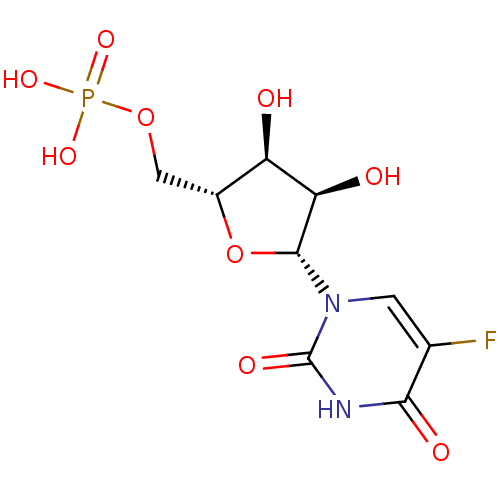
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bello, AM; Konforte, D; Poduch, E; Furlonger, C; Wei, L; Liu, Y; Lewis, M; Pai, EF; Paige, CJ; Kotra, LP Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents. J Med Chem52:1648-58 (2009) [PubMed] Article
Bello, AM; Konforte, D; Poduch, E; Furlonger, C; Wei, L; Liu, Y; Lewis, M; Pai, EF; Paige, CJ; Kotra, LP Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents. J Med Chem52:1648-58 (2009) [PubMed] Article