| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor gamma |
|---|
| Ligand | BDBM29868 |
|---|
| Substrate/Competitor | [3H]rosiglitazone |
|---|
| Meas. Tech. | Scintillation Proximity Assay (Ki) and Cell-Based Transcription Assay (EC50) |
|---|
| Ki | 300±n/a nM |
|---|
| EC50 | 3000±n/a nM |
|---|
| Citation |  Connors, RV; Wang, Z; Harrison, M; Zhang, A; Wanska, M; Hiscock, S; Fox, B; Dore, M; Labelle, M; Sudom, A; Johnstone, S; Liu, J; Walker, NP; Chai, A; Siegler, K; Li, Y; Coward, P Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma. Bioorg Med Chem Lett19:3550-4 (2009) [PubMed] Article Connors, RV; Wang, Z; Harrison, M; Zhang, A; Wanska, M; Hiscock, S; Fox, B; Dore, M; Labelle, M; Sudom, A; Johnstone, S; Liu, J; Walker, NP; Chai, A; Siegler, K; Li, Y; Coward, P Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma. Bioorg Med Chem Lett19:3550-4 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor gamma |
|---|
| Name: | Peroxisome proliferator-activated receptor gamma |
|---|
| Synonyms: | NR1C3 | Nuclear receptor subfamily 1 group C member 3 | PPAR-gamma | PPARG | PPARG_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor gamma (PPAR gamma) | Peroxisome proliferator-activated receptor gamma (PPARG) | Peroxisome proliferator-activated receptor gamma (PPARγ) | Peroxisome proliferator-activated receptor gamma/Nuclear receptor corepressor 2 | peroxisome proliferator-activated receptor gamma isoform 2 |
|---|
| Type: | Nuclear Receptor |
|---|
| Mol. Mass.: | 57613.46 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P37231 |
|---|
| Residue: | 505 |
|---|
| Sequence: | MGETLGDSPIDPESDSFTDTLSANISQEMTMVDTEMPFWPTNFGISSVDLSVMEDHSHSF
DIKPFTTVDFSSISTPHYEDIPFTRTDPVVADYKYDLKLQEYQSAIKVEPASPPYYSEKT
QLYNKPHEEPSNSLMAIECRVCGDKASGFHYGVHACEGCKGFFRRTIRLKLIYDRCDLNC
RIHKKSRNKCQYCRFQKCLAVGMSHNAIRFGRMPQAEKEKLLAEISSDIDQLNPESADLR
ALAKHLYDSYIKSFPLTKAKARAILTGKTTDKSPFVIYDMNSLMMGEDKIKFKHITPLQE
QSKEVAIRIFQGCQFRSVEAVQEITEYAKSIPGFVNLDLNDQVTLLKYGVHEIIYTMLAS
LMNKDGVLISEGQGFMTREFLKSLRKPFGDFMEPKFEFAVKFNALELDDSDLAIFIAVII
LSGDRPGLLNVKPIEDIQDNLLQALELQLKLNHPESSQLFAKLLQKMTDLRQIVTEHVQL
LQVIKKTETDMSLHPLLQEIYKDLY
|
|
|
|---|
| BDBM29868 |
|---|
| [3H]rosiglitazone |
|---|
| Name | BDBM29868 |
|---|
| Synonyms: | alkynyl ether, 10 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H23F3O5S |
|---|
| Mol. Mass. | 516.529 |
|---|
| SMILES | Cc1c(C)c(Sc2ccc(COc3ccc(cc3)C(F)(F)F)cc2OCC#C)ccc1OCC(O)=O |
|---|
| Structure | 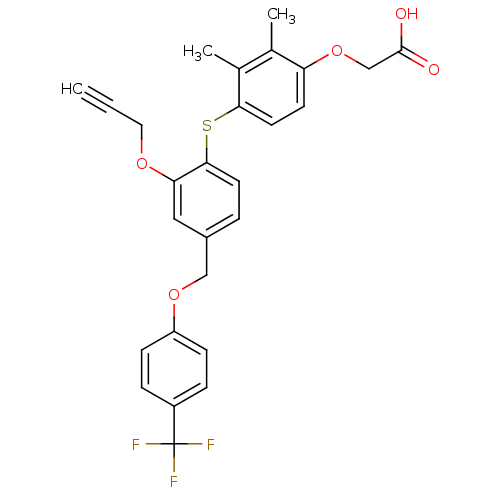
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Connors, RV; Wang, Z; Harrison, M; Zhang, A; Wanska, M; Hiscock, S; Fox, B; Dore, M; Labelle, M; Sudom, A; Johnstone, S; Liu, J; Walker, NP; Chai, A; Siegler, K; Li, Y; Coward, P Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma. Bioorg Med Chem Lett19:3550-4 (2009) [PubMed] Article
Connors, RV; Wang, Z; Harrison, M; Zhang, A; Wanska, M; Hiscock, S; Fox, B; Dore, M; Labelle, M; Sudom, A; Johnstone, S; Liu, J; Walker, NP; Chai, A; Siegler, K; Li, Y; Coward, P Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma. Bioorg Med Chem Lett19:3550-4 (2009) [PubMed] Article