| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA polymerase III subunit tau |
|---|
| Ligand | BDBM36317 |
|---|
| Substrate/Competitor | BDBM50118233 |
|---|
| Meas. Tech. | colorimetric ATPase assay |
|---|
| pH | 7.5±0 |
|---|
| Temperature | 310.15±0 K |
|---|
| Ki | 3.00e+4± 1.57e+4 nM |
|---|
| Citation |  Eng, K; Scouten-Ponticelli, SK; Sutton, M; Berdis, A Selective Inhibition of DNA Replicase Assembly by a Non-natural Nucleotide: Exploiting the Structural Diversity of ATP-Binding Sites ACS Chem Biol5:183-94 (2010) [PubMed] Article Eng, K; Scouten-Ponticelli, SK; Sutton, M; Berdis, A Selective Inhibition of DNA Replicase Assembly by a Non-natural Nucleotide: Exploiting the Structural Diversity of ATP-Binding Sites ACS Chem Biol5:183-94 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| DNA polymerase III subunit tau |
|---|
| Name: | DNA polymerase III subunit tau |
|---|
| Synonyms: | ATP-dependent clamp loader gamma -complex | DNA polymerase III subunit gamma | DPO3X_ECOLI | dnaX | dnaZ | dnaZX |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 71139.93 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | P06710 |
|---|
| Residue: | 643 |
|---|
| Sequence: | MSYQVLARKWRPQTFADVVGQEHVLTALANGLSLGRIHHAYLFSGTRGVGKTSIARLLAK
GLNCETGITATPCGVCDNCREIEQGRFVDLIEIDAASRTKVEDTRDLLDNVQYAPARGRF
KVYLIDEVHMLSRHSFNALLKTLEEPPEHVKFLLATTDPQKLPVTILSRCLQFHLKALDV
EQIRHQLEHILNEEHIAHEPRALQLLARAAEGSLRDALSLTDQAIASGDGQVSTQAVSAM
LGTLDDDQALSLVEAMVEANGERVMALINEAAARGIEWEALLVEMLGLLHRIAMVQLSPA
ALGNDMAAIELRMRELARTIPPTDIQLYYQTLLIGRKELPYAPDRRMGVEMTLLRALAFH
PRMPLPEPEVPRQSFAPVAPTAVMTPTQVPPQPQSAPQQAPTVPLPETTSQVLAARQQLQ
RVQGATKAKKSEPAAATRARPVNNAALERLASVTDRVQARPVPSALEKAPAKKEAYRWKA
TTPVMQQKEVVATPKALKKALEHEKTPELAAKLAAEAIERDPWAAQVSQLSLPKLVEQVA
LNAWKEESDNAVCLHLRSSQRHLNNRGAQQKLAEALSMLKGSTVELTIVEDDNPAVRTPL
EWRQAIYEEKLAQARESIIADNNIQTLRRFFDAELDEESIRPI
|
|
|
|---|
| BDBM36317 |
|---|
| BDBM50118233 |
|---|
| Name | BDBM36317 |
|---|
| Synonyms: | 5-ethylene-indolyl- 2'-deoxyriboside triphosphate | 5-ethylene-indolyl- 2=-deoxyriboside triphosphate | d5-EyITP |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H20NO12P3 |
|---|
| Mol. Mass. | 499.2401 |
|---|
| SMILES | [H][C@]1(O)C[C@@]([H])(O[C@]1([H])COP(O)(=O)OP(O)(=O)OP(O)(O)=O)n1ccc2cc(C=C)ccc12 |
|---|
| Structure | 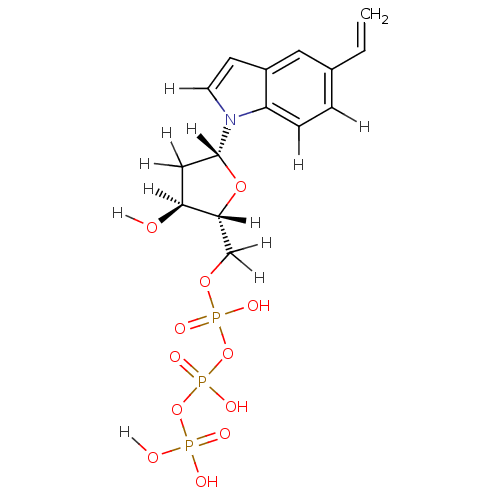
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Eng, K; Scouten-Ponticelli, SK; Sutton, M; Berdis, A Selective Inhibition of DNA Replicase Assembly by a Non-natural Nucleotide: Exploiting the Structural Diversity of ATP-Binding Sites ACS Chem Biol5:183-94 (2010) [PubMed] Article
Eng, K; Scouten-Ponticelli, SK; Sutton, M; Berdis, A Selective Inhibition of DNA Replicase Assembly by a Non-natural Nucleotide: Exploiting the Structural Diversity of ATP-Binding Sites ACS Chem Biol5:183-94 (2010) [PubMed] Article