| Reaction Details |
|---|
 | Report a problem with these data |
| Target | HIV-1 protease |
|---|
| Ligand | BDBM36652 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Protease Inhibtion Assay |
|---|
| pH | 5.5±0 |
|---|
| Ki | 0.34±0.0 nM |
|---|
| Citation |  Rodgers, JD; Lam, PY; Johnson, BL; Wang, H; Li, R; Ru, Y; Ko, SS; Seitz, SP; Trainor, GL; Anderson, PS; Klabe, RM; Bacheler, LT; Cordova, B; Garber, S; Reid, C; Wright, MR; Chang, CH; Erickson-Viitanen, S Design and selection of DMP 850 and DMP 851: the next generation of cyclic urea HIV protease inhibitors. Chem Biol5:597-608 (1998) [PubMed] Article Rodgers, JD; Lam, PY; Johnson, BL; Wang, H; Li, R; Ru, Y; Ko, SS; Seitz, SP; Trainor, GL; Anderson, PS; Klabe, RM; Bacheler, LT; Cordova, B; Garber, S; Reid, C; Wright, MR; Chang, CH; Erickson-Viitanen, S Design and selection of DMP 850 and DMP 851: the next generation of cyclic urea HIV protease inhibitors. Chem Biol5:597-608 (1998) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| HIV-1 protease |
|---|
| Name: | HIV-1 protease |
|---|
| Synonyms: | HIV-1 protease wild type |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 10757.68 |
|---|
| Organism: | Human immunodeficiency virus |
|---|
| Description: | O90785 |
|---|
| Residue: | 99 |
|---|
| Sequence: | PQITLWQRPLVTVKIGGQLREALLDTGADDTVLEDINLPGKWKPKMIGGIGGFIKVKQYE
QVLIEICGKKAIGTVLVGPTPVNIIGRNMLTQIGCTLNF
|
|
|
|---|
| BDBM36652 |
|---|
| n/a |
|---|
| Name | BDBM36652 |
|---|
| Synonyms: | 3-alkylaminoindazole cyclic urea, (c-PrCH2-) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C41H44N8O3 |
|---|
| Mol. Mass. | 696.8399 |
|---|
| SMILES | O[C@@H]1[C@@H](O)[C@@H](Cc2ccccc2)N(Cc2ccc3[nH]nc(NC4CC4)c3c2)C(=O)N(Cc2ccc3[nH]nc(NC4CC4)c3c2)[C@@H]1Cc1ccccc1 |r| |
|---|
| Structure | 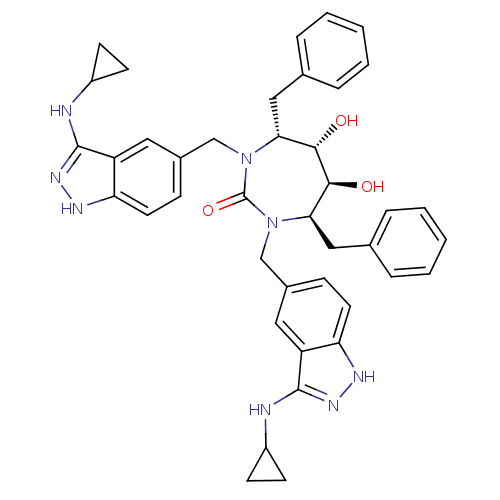
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rodgers, JD; Lam, PY; Johnson, BL; Wang, H; Li, R; Ru, Y; Ko, SS; Seitz, SP; Trainor, GL; Anderson, PS; Klabe, RM; Bacheler, LT; Cordova, B; Garber, S; Reid, C; Wright, MR; Chang, CH; Erickson-Viitanen, S Design and selection of DMP 850 and DMP 851: the next generation of cyclic urea HIV protease inhibitors. Chem Biol5:597-608 (1998) [PubMed] Article
Rodgers, JD; Lam, PY; Johnson, BL; Wang, H; Li, R; Ru, Y; Ko, SS; Seitz, SP; Trainor, GL; Anderson, PS; Klabe, RM; Bacheler, LT; Cordova, B; Garber, S; Reid, C; Wright, MR; Chang, CH; Erickson-Viitanen, S Design and selection of DMP 850 and DMP 851: the next generation of cyclic urea HIV protease inhibitors. Chem Biol5:597-608 (1998) [PubMed] Article