| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase BTK |
|---|
| Ligand | BDBM4779 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Kinase Inhibitor Selectivity Profiling Assay |
|---|
| Kd | 1600±n/a nM |
|---|
| Citation |  PubChem, PC Kinase Inhibitor Selectivity Profiling Assay PubChem Bioassay(2008)[AID] PubChem, PC Kinase Inhibitor Selectivity Profiling Assay PubChem Bioassay(2008)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Tyrosine-protein kinase BTK |
|---|
| Name: | Tyrosine-protein kinase BTK |
|---|
| Synonyms: | AGMX1 | ATK | Agammaglobulinaemia tyrosine kinase | Agammaglobulinemia tyrosine kinase | B cell progenitor kinase | B-cell progenitor kinase | BPK | BTK | BTK_HUMAN | Bruton tyrosine kinase | Tyrosine Kinase BTK | Tyrosine-protein kinase (BTK) | Tyrosine-protein kinase BTK (BTK) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 76289.95 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q06187 |
|---|
| Residue: | 659 |
|---|
| Sequence: | MAAVILESIFLKRSQQKKKTSPLNFKKRLFLLTVHKLSYYEYDFERGRRGSKKGSIDVEK
ITCVETVVPEKNPPPERQIPRRGEESSEMEQISIIERFPYPFQVVYDEGPLYVFSPTEEL
RKRWIHQLKNVIRYNSDLVQKYHPCFWIDGQYLCCSQTAKNAMGCQILENRNGSLKPGSS
HRKTKKPLPPTPEEDQILKKPLPPEPAAAPVSTSELKKVVALYDYMPMNANDLQLRKGDE
YFILEESNLPWWRARDKNGQEGYIPSNYVTEAEDSIEMYEWYSKHMTRSQAEQLLKQEGK
EGGFIVRDSSKAGKYTVSVFAKSTGDPQGVIRHYVVCSTPQSQYYLAEKHLFSTIPELIN
YHQHNSAGLISRLKYPVSQQNKNAPSTAGLGYGSWEIDPKDLTFLKELGTGQFGVVKYGK
WRGQYDVAIKMIKEGSMSEDEFIEEAKVMMNLSHEKLVQLYGVCTKQRPIFIITEYMANG
CLLNYLREMRHRFQTQQLLEMCKDVCEAMEYLESKQFLHRDLAARNCLVNDQGVVKVSDF
GLSRYVLDDEYTSSVGSKFPVRWSPPEVLMYSKFSSKSDIWAFGVLMWEIYSLGKMPYER
FTNSETAEHIAQGLRLYRPHLASEKVYTIMYSCWHEKADERPTFKILLSNILDVMDEES
|
|
|
|---|
| BDBM4779 |
|---|
| n/a |
|---|
| Name | BDBM4779 |
|---|
| Synonyms: | CHEMBL31965 | CHEMBL545315 | CI-1033 | Canertinib | N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[3-(morpholin-4-yl)propoxy]quinazolin-6-yl}prop-2-enamide | PD0183805 | US10507209, Compound CI-1033 | US9730934, CI-1033 | cid_156414 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H25ClFN5O3 |
|---|
| Mol. Mass. | 485.938 |
|---|
| SMILES | Fc1ccc(Nc2ncnc3cc(OCCCN4CCOCC4)c(NC(=O)C=C)cc23)cc1Cl |
|---|
| Structure | 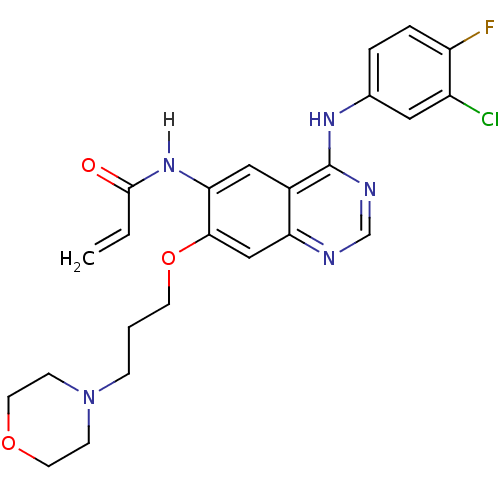
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Kinase Inhibitor Selectivity Profiling Assay PubChem Bioassay(2008)[AID]
PubChem, PC Kinase Inhibitor Selectivity Profiling Assay PubChem Bioassay(2008)[AID]