| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-lactamase |
|---|
| Ligand | BDBM39823 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assay |
|---|
| pH | 7±0 |
|---|
| Ki | 7.0e+2±n/a nM |
|---|
| Citation |  Tondi, D; Powers, RA; Caselli, E; Negri, MC; Blázquez, J; Costi, MP; Shoichet, BK Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase. Chem Biol8:593-611 (2001) [PubMed] Article Tondi, D; Powers, RA; Caselli, E; Negri, MC; Blázquez, J; Costi, MP; Shoichet, BK Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase. Chem Biol8:593-611 (2001) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Beta-lactamase |
|---|
| Name: | Beta-lactamase |
|---|
| Synonyms: | AMPC_ECOLI | Beta-lactamase | Beta-lactamase (AmpC) | Beta-lactamase AmpC | Cephalosporinase | Escherichia coli K-12 | ampA | ampC |
|---|
| Type: | Protien |
|---|
| Mol. Mass.: | 41561.62 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | P00811 |
|---|
| Residue: | 377 |
|---|
| Sequence: | MFKTTLCALLITASCSTFAAPQQINDIVHRTITPLIEQQKIPGMAVAVIYQGKPYYFTWG
YADIAKKQPVTQQTLFELGSVSKTFTGVLGGDAIARGEIKLSDPTTKYWPELTAKQWNGI
TLLHLATYTAGGLPLQVPDEVKSSSDLLRFYQNWQPAWAPGTQRLYANSSIGLFGALAVK
PSGLSFEQAMQTRVFQPLKLNHTWINVPPAEEKNYAWGYREGKAVHVSPGALDAEAYGVK
STIEDMARWVQSNLKPLDINEKTLQQGIQLAQSRYWQTGDMYQGLGWEMLDWPVNPDSII
NGSDNKIALAARPVKAITPPTPAVRASWVHKTGATGGFGSYVAFIPEKELGIVMLANKNY
PNPARVDAAWQILNALQ
|
|
|
|---|
| BDBM39823 |
|---|
| n/a |
|---|
| Name | BDBM39823 |
|---|
| Synonyms: | Amide and sulfonamide derivatives, 7 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H11BF3NO5S |
|---|
| Mol. Mass. | 361.101 |
|---|
| SMILES | OB(O)c1cccc(NS(=O)(=O)c2ccc(OC(F)(F)F)cc2)c1 |
|---|
| Structure | 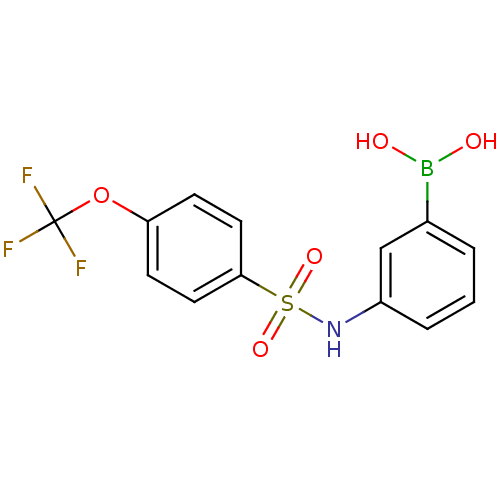
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tondi, D; Powers, RA; Caselli, E; Negri, MC; Blázquez, J; Costi, MP; Shoichet, BK Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase. Chem Biol8:593-611 (2001) [PubMed] Article
Tondi, D; Powers, RA; Caselli, E; Negri, MC; Blázquez, J; Costi, MP; Shoichet, BK Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase. Chem Biol8:593-611 (2001) [PubMed] Article