| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phosphotransferase |
|---|
| Ligand | BDBM43835 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assay |
|---|
| Temperature | 298.15±0 K |
|---|
| Ki | 2.8e+3± 1e+2 nM |
|---|
| IC50 | 500000±0.0 nM |
|---|
| Citation |  Willson, M; Sanejouand, YH; Perie, J; Hannaert, V; Opperdoes, F Sequencing, modeling, and selective inhibition of Trypanosoma brucei hexokinase. Chem Biol9:839-47 (2002) [PubMed] Article Willson, M; Sanejouand, YH; Perie, J; Hannaert, V; Opperdoes, F Sequencing, modeling, and selective inhibition of Trypanosoma brucei hexokinase. Chem Biol9:839-47 (2002) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Phosphotransferase |
|---|
| Name: | Phosphotransferase |
|---|
| Synonyms: | Hexokinase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 51306.14 |
|---|
| Organism: | Trypanosoma brucei |
|---|
| Description: | Q38C42 |
|---|
| Residue: | 471 |
|---|
| Sequence: | MSRRLNNILEHISIQGNDGETVRAVKRDVAMAALTNQFTMSVESMRQIMTYLLYEMVEGL
EGRESTVRMLPSYVYKADPKRATGVFYALDLGGTNFRVLRVACKEGAVVDSSTSAFKIPK
YALEGNATDLFGFIASNVKKTMETRAPEDLNRTVPLGFTFSFPVEQTKVNRGVLIRWTKG
FSTKGVQGNDVIALLQAAFGRVSLKVNVVALCNDTVGTLISHYFKDPEVQVGVIIGTGSN
ACYFETASAVTKDPAVAARGSALTPINMESGNFDSKYRFVLPTTKFDLDIDDASLNKGQQ
ALEKMISGMYLGEIARRVIVHLSSINCLPAALQTALGNRGSFESRFAGMISADRMPGLQF
TRSTIQKVCGVDVQSIEDLRIIRDVCRLVRGRAAQLSASFCCAPLVKTQTQGRATIAIDG
SVFEKIPSFRRVLQDNINRILGPECDVRAVLAKDGSGIGAAFISAMVVNDK
|
|
|
|---|
| BDBM43835 |
|---|
| n/a |
|---|
| Name | BDBM43835 |
|---|
| Synonyms: | Glucosamine derivative, 8 | MLS000582359 | N-cyclohexyl-5-nitro-2-(1-piperidinyl)benzamide | N-cyclohexyl-5-nitro-2-piperidin-1-yl-benzamide | N-cyclohexyl-5-nitro-2-piperidin-1-ylbenzamide | N-cyclohexyl-5-nitro-2-piperidino-benzamide | SMR000200900 | cid_2967968 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H25N3O3 |
|---|
| Mol. Mass. | 331.4094 |
|---|
| SMILES | [O-][N+](=O)c1ccc(N2CCCCC2)c(c1)C(=O)NC1CCCCC1 |
|---|
| Structure | 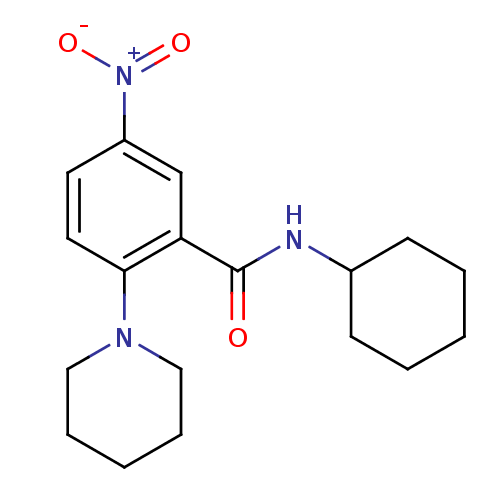
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Willson, M; Sanejouand, YH; Perie, J; Hannaert, V; Opperdoes, F Sequencing, modeling, and selective inhibition of Trypanosoma brucei hexokinase. Chem Biol9:839-47 (2002) [PubMed] Article
Willson, M; Sanejouand, YH; Perie, J; Hannaert, V; Opperdoes, F Sequencing, modeling, and selective inhibition of Trypanosoma brucei hexokinase. Chem Biol9:839-47 (2002) [PubMed] Article