| Reaction Details |
|---|
 | Report a problem with these data |
| Target | HIV-1 protease |
|---|
| Ligand | BDBM586093 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assay |
|---|
| pH | 5.6±0 |
|---|
| Temperature | 310.15±0 K |
|---|
| IC50 | >50±0.5 nM |
|---|
| Citation |  Brik, A; Lin, YC; Elder, J; Wong, CH A quick diversity-oriented amide-forming reaction to optimize P-subsite residues of HIV protease inhibitors. Chem Biol9:891-6 (2002) [PubMed] Article Brik, A; Lin, YC; Elder, J; Wong, CH A quick diversity-oriented amide-forming reaction to optimize P-subsite residues of HIV protease inhibitors. Chem Biol9:891-6 (2002) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| HIV-1 protease |
|---|
| Name: | HIV-1 protease |
|---|
| Synonyms: | HIV-1 protease wild type |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 10757.68 |
|---|
| Organism: | Human immunodeficiency virus |
|---|
| Description: | O90785 |
|---|
| Residue: | 99 |
|---|
| Sequence: | PQITLWQRPLVTVKIGGQLREALLDTGADDTVLEDINLPGKWKPKMIGGIGGFIKVKQYE
QVLIEICGKKAIGTVLVGPTPVNIIGRNMLTQIGCTLNF
|
|
|
|---|
| BDBM586093 |
|---|
| n/a |
|---|
| Name | BDBM586093 |
|---|
| Synonyms: | P3-P3' Entry 11 |
|---|
| Type | n/a |
|---|
| Emp. Form. | C52H50Cl2F6N4O8 |
|---|
| Mol. Mass. | 1043.872 |
|---|
| SMILES | CC(C)[C@H](NC(=O)c1ccc(o1)-c1cc(ccc1Cl)C(F)(F)F)C(=O)N[C@@H](Cc1ccccc1)[C@@H](O)[C@H](O)[C@H](Cc1ccccc1)NC(=O)[C@@H](NC(=O)c1ccc(o1)-c1cc(ccc1Cl)C(F)(F)F)C(C)C |
|---|
| Structure | 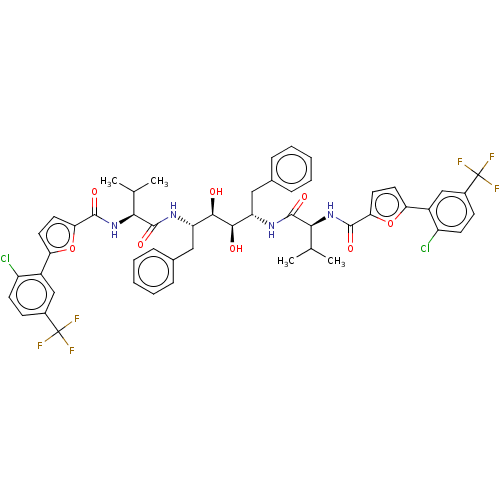
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Brik, A; Lin, YC; Elder, J; Wong, CH A quick diversity-oriented amide-forming reaction to optimize P-subsite residues of HIV protease inhibitors. Chem Biol9:891-6 (2002) [PubMed] Article
Brik, A; Lin, YC; Elder, J; Wong, CH A quick diversity-oriented amide-forming reaction to optimize P-subsite residues of HIV protease inhibitors. Chem Biol9:891-6 (2002) [PubMed] Article