| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glycylpeptide N-tetradecanoyltransferase 1 |
|---|
| Ligand | BDBM96235 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assay |
|---|
| IC50 | 1.94e+5±n/a nM |
|---|
| Citation |  Sogabe, S; Masubuchi, M; Sakata, K; Fukami, TA; Morikami, K; Shiratori, Y; Ebiike, H; Kawasaki, K; Aoki, Y; Shimma, N; D'Arcy, A; Winkler, FK; Banner, DW; Ohtsuka, T Crystal structures of Candida albicans N-myristoyltransferase with two distinct inhibitors. Chem Biol9:1119-28 (2002) [PubMed] Article Sogabe, S; Masubuchi, M; Sakata, K; Fukami, TA; Morikami, K; Shiratori, Y; Ebiike, H; Kawasaki, K; Aoki, Y; Shimma, N; D'Arcy, A; Winkler, FK; Banner, DW; Ohtsuka, T Crystal structures of Candida albicans N-myristoyltransferase with two distinct inhibitors. Chem Biol9:1119-28 (2002) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glycylpeptide N-tetradecanoyltransferase 1 |
|---|
| Name: | Glycylpeptide N-tetradecanoyltransferase 1 |
|---|
| Synonyms: | Myristoyl-CoA:protein N-myristoyltransferase (Nmt) | N-myristoyltransferase (NMT1) | NMT | NMT1 | NMT1_HUMAN | Peptide N-myristoyltransferase | Peptide N-myristoyltransferase 1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 56814.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P30419 |
|---|
| Residue: | 496 |
|---|
| Sequence: | MADESETAVKPPAPPLPQMMEGNGNGHEHCSDCENEEDNSYNRGGLSPANDTGAKKKKKK
QKKKKEKGSETDSAQDQPVKMNSLPAERIQEIQKAIELFSVGQGPAKTMEEASKRSYQFW
DTQPVPKLGEVVNTHGPVEPDKDNIRQEPYTLPQGFTWDALDLGDRGVLKELYTLLNENY
VEDDDNMFRFDYSPEFLLWALRPPGWLPQWHCGVRVVSSRKLVGFISAIPANIHIYDTEK
KMVEINFLCVHKKLRSKRVAPVLIREITRRVHLEGIFQAVYTAGVVLPKPVGTCRYWHRS
LNPRKLIEVKFSHLSRNMTMQRTMKLYRLPETPKTAGLRPMETKDIPVVHQLLTRYLKQF
HLTPVMSQEEVEHWFYPQENIIDTFVVENANGEVTDFLSFYTLPSTIMNHPTHKSLKAAY
SFYNVHTQTPLLDLMSDALVLAKMKGFDVFNALDLMENKTFLEKLKFGIGDGNLQYYLYN
WKCPSMGAEKVGLVLQ
|
|
|
|---|
| BDBM96235 |
|---|
| n/a |
|---|
| Name | BDBM96235 |
|---|
| Synonyms: | 4-[2-hydroxy-3-(isopropylamino)propoxy]-3-methyl-coumarilic acid ethyl ester;hydrochloride | 4-[2-hydroxy-3-(propan-2-ylamino)propoxy]-3-methyl-2-benzofurancarboxylic acid ethyl ester;hydrochloride | CHEMBL53849 | MLS000039218 | Non peptidic, 1 | SMR000036354 | cid_6602742 | ethyl 3-methyl-4-[2-oxidanyl-3-(propan-2-ylamino)propoxy]-1-benzofuran-2-carboxylate;hydrochloride | ethyl 4-[2-hydroxy-3-(propan-2-ylamino)propoxy]-3-methyl-1-benzofuran-2-carboxylate;hydrochloride |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H25NO5 |
|---|
| Mol. Mass. | 335.3948 |
|---|
| SMILES | CCOC(=O)c1oc2cccc(OCC(O)CNC(C)C)c2c1C |
|---|
| Structure | 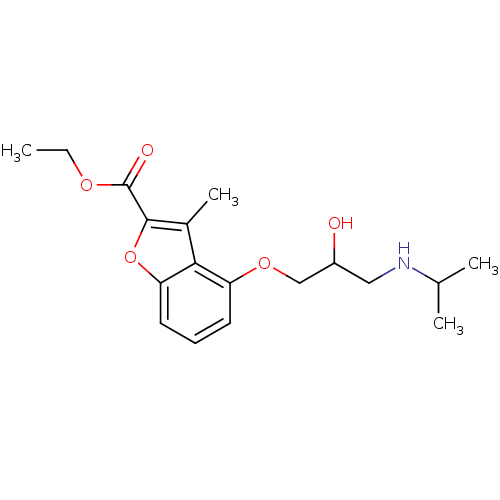
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sogabe, S; Masubuchi, M; Sakata, K; Fukami, TA; Morikami, K; Shiratori, Y; Ebiike, H; Kawasaki, K; Aoki, Y; Shimma, N; D'Arcy, A; Winkler, FK; Banner, DW; Ohtsuka, T Crystal structures of Candida albicans N-myristoyltransferase with two distinct inhibitors. Chem Biol9:1119-28 (2002) [PubMed] Article
Sogabe, S; Masubuchi, M; Sakata, K; Fukami, TA; Morikami, K; Shiratori, Y; Ebiike, H; Kawasaki, K; Aoki, Y; Shimma, N; D'Arcy, A; Winkler, FK; Banner, DW; Ohtsuka, T Crystal structures of Candida albicans N-myristoyltransferase with two distinct inhibitors. Chem Biol9:1119-28 (2002) [PubMed] Article